
- Jun. 25, 2025
- Home
- About Us
- Editorial Board
- Instruction
- Subscription
- Advertisement
- Contact Us
- Chinese
- RSS

Chinese Journal of Magnetic Resonance ›› 2024, Vol. 41 ›› Issue (1): 1-8.doi: 10.11938/cjmr20233069cstr: 32225.14.cjmr20233069
• Articles • Previous Articles Next Articles
WANG Huan1,TAO Zhiqing2,3,JIANG Guosheng1,ZHANG Xu3,WANG Guan3( ),HE Lichun2,3(
),HE Lichun2,3( ),LIU Maili3
),LIU Maili3
Received:2023-05-16
Published:2024-03-05
Online:2023-06-30
Contact:
Tel: 86-18602761433, E-mail: CLC Number:
WANG Huan, TAO Zhiqing, JIANG Guosheng, ZHANG Xu, WANG Guan, HE Lichun, LIU Maili. In situ Investigation of HdeA in Bacterial Outer Membrane Vesicles Using NMR Spectroscopy[J]. Chinese Journal of Magnetic Resonance, 2024, 41(1): 1-8.
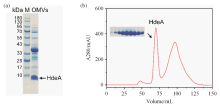
Fig. 2
(a) SDS-PAGE gel plot of HdeA-OMVs samples, the arrow marked band (~10 kDa) is HdeA, and the remaining bands are pericytoplasmic proteins or outer membrane proteins in OMVs; (b) Liquid chromatogram of the HdeA from medium supernatant separated after size exclusion chromatography, in which the peak of the dimer HdeA is located at about 68 mL
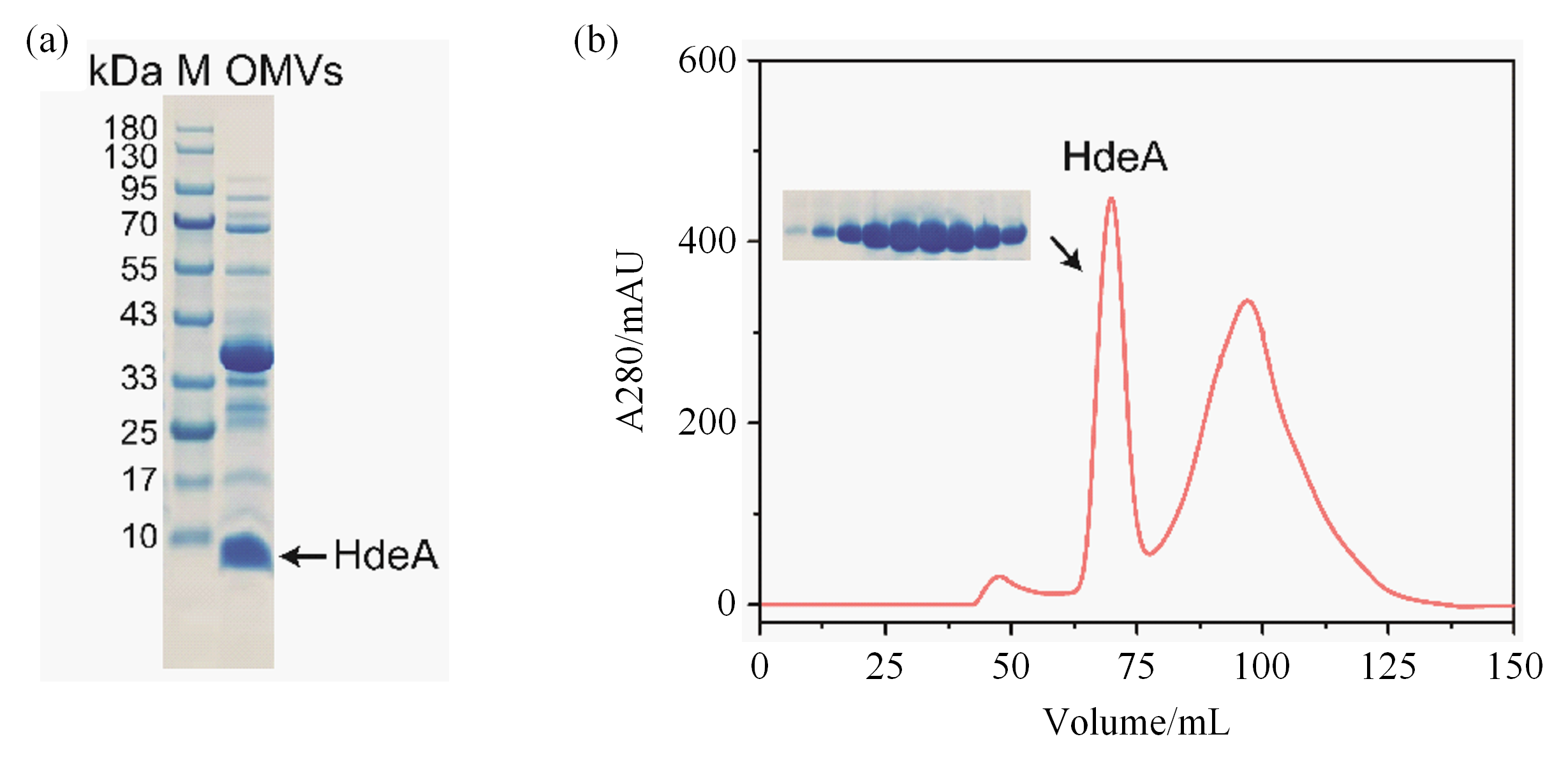
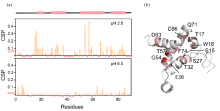
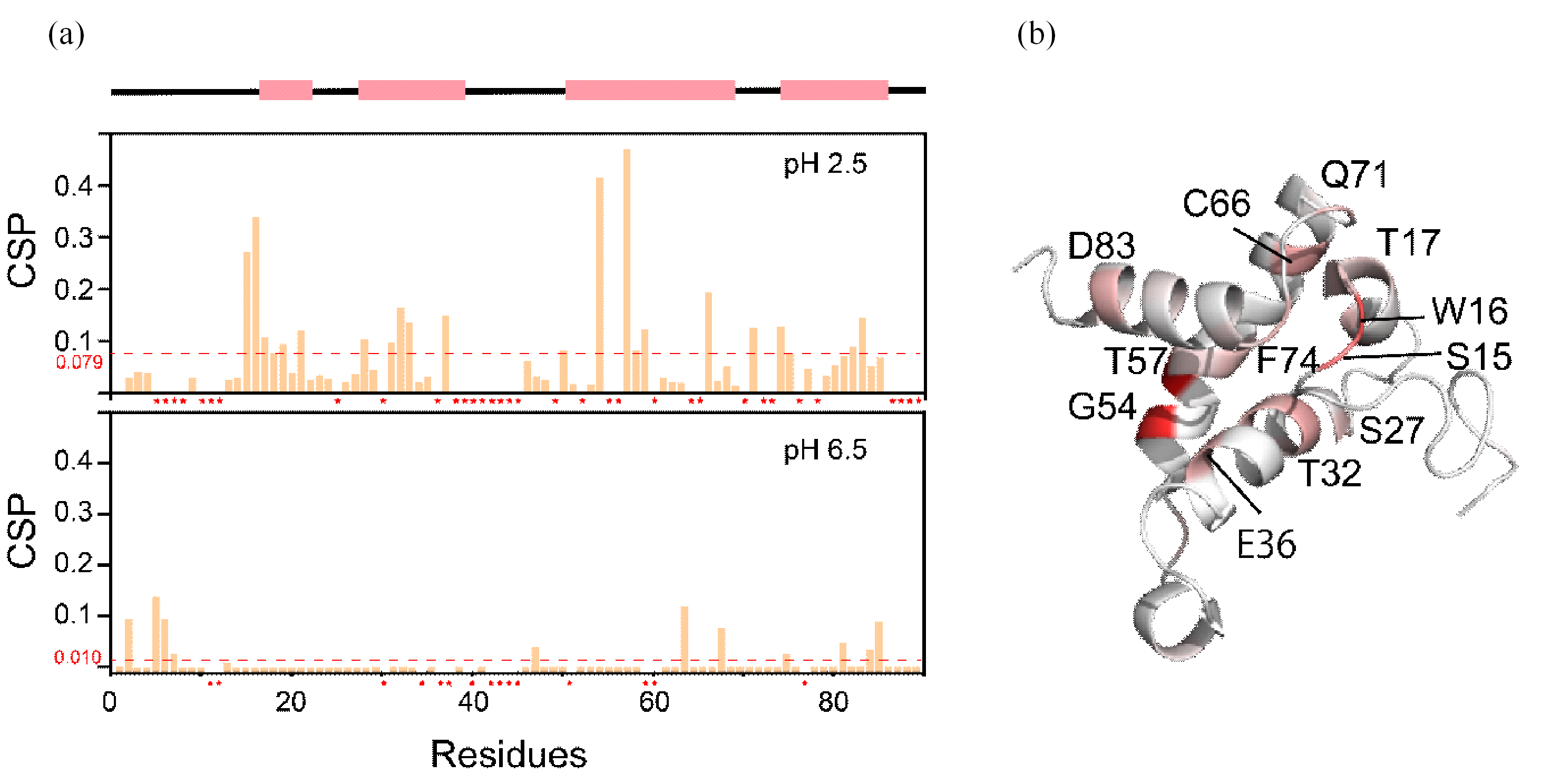
Fig. 5
The CSP of residues of HdeA in OMVs. (a) Chemical shift perturbation difference of each residue of HdeA in the OMVs environment and dilute solution at pH 2.5 and pH 6.5, where * indicates that no credible CSP data could be obtained for this residue due to peak overlap; (b) Monomer structure of HdeA, in which residues with large variation in CSP values are marked in red
| [1] |
CHRISTOPH W, ANDREAS P. Protein folding in the periplasm of Escherichia coli[J]. Mol Microbiol, 1994, 12: 685-692.
doi: 10.1111/mmi.1994.12.issue-5 |
| [2] |
HONG W Z, WU Y E, FU X M, et al. Chaperone-dependent mechanisms for acid resistance in enteric bacteria[J]. Trends Microbiol, 2012, 20(7): 328-335.
doi: 10.1016/j.tim.2012.03.001 pmid: 22459131 |
| [3] |
YU X C, HU Y F, DING J, et al. Structural basis and mechanism of the unfolding-induced activation of HdeA, a bacterial acid response chaperone[J]. J Biol Chem, 2019, 294(9): 3192-3206.
doi: 10.1074/jbc.RA118.006398 |
| [4] |
GARRISON M A, CROWHURST K A. NMR-monitored titration of acid-stress bacterial chaperone HdeA reveals that Asp and Glu charge neutralization produces a loosened dimer structure in preparation for protein unfolding and chaperone activation[J]. Protein Sci, 2014, 23(2): 167-178.
doi: 10.1002/pro.2402 pmid: 24375557 |
| [5] | ZHAN J H, HU Q, ZHU Q J, et al. Track the conformational change of unlabeled yeast cytochrome c in cell homogenate using NMR[J]. Chinese J Magn Reson, 2023, 40(1): 22-29. |
| 占建华, 胡琴, 朱勤俊, 等. 基于磁共振的胞浆中无标记酵母细胞色素c构象变化追踪[J]. 波谱学杂志, 2023, 40(1): 22-29. | |
| [6] |
XING C Y, CHENGFENG Y, JIENV D, et al. Characterizations of the interactions between Escherichia coli periplasmic chaperone HdeA and its native substrates during acid stress[J]. Biochemistry, 2017, 56 (43): 5748-5757
doi: 10.1021/acs.biochem.7b00724 |
| [7] |
SALMON L, STULL F, SAYLE S, et al. The mechanism of HdeA unfolding and chaperone activation[J]. J Mol Biol, 2018, 430(1): 33-40.
doi: S0022-2836(17)30540-5 pmid: 29138002 |
| [8] |
ZHAI Z N, WU Q, ZHENG W W, et al. Roles of structural plasticity in chaperone HdeA activity are revealed by 19F NMR[J]. Chem Sci, 2016, 7: 2222.
doi: 10.1039/C5SC04297F |
| [9] |
WANG G, YU G J, GAO D W. Protein conformational exchanges modulated by the environment of outer membrane vesicles[J]. J Phys Chem Lett, 2023, 14 (11): 2772-2777.
doi: 10.1021/acs.jpclett.3c00152 pmid: 36897994 |
| [10] |
THOMA J, MANIOGLU S, KALBERMATTER D, et al. Protein-enriched outer membrane vesicles as a native platform for outer membrane protein studies[J]. Comm Biol, 2018, 1: 23.
doi: 10.1038/s42003-018-0027-5 |
| [11] |
NIKAIDO H, VAARA M. Molecular basis of bacterial outer membrane permeability[J]. Microbiol Rev, 1985, 49(1): 1-32.
doi: 10.1128/mr.49.1.1-32.1985 pmid: 2580220 |
| [12] |
SCHIRMER T. General and specific porins from bacterial outer membranes[J]. J Struct Biol, 1998, 121(2): 101-109.
pmid: 9615433 |
| [13] |
KOEBNIK R, LOCHER K P, VAN GELDER P. Structure and function of bacterial outer membrane proteins: barrels in a nutshell[J]. Mol Microbiol, 2000, 37(2): 239-253.
doi: 10.1046/j.1365-2958.2000.01983.x pmid: 10931321 |
| [14] |
JOHANNES T, BJÖRN M B. High-resolution in situ NMR spectroscopy of bacterial envelope proteins in outer membrane vesicles[J]. Biochemistry, 2020, 59(17): 1656-1660.
doi: 10.1021/acs.biochem.9b01123 |
| [15] |
VRANKEN W F, BOUCHER W, STEVENS T J, et al. The CCPN data model for NMR spectroscopy: development of a software pipeline[J]. Proteins, 2005, 59(4): 687-696.
doi: 10.1002/prot.v59:4 |
| [16] | HOEKSTRA D, VAN DER LAAN J W, DE LEIJ L, et al. Release of outer membrane fragments from normally growing Escherichia coli[J]. Biochim Biophys Acta, 1976, 455(3): 889-899. |
| [17] | MUG-OPSTELTEN D, WITHOLT B. Preferential release of new outer membrane fragments by exponentially growing Escherichia coli[J]. Biochim Biophys Acta, 1978, 508(2): 287-295. |
| [18] |
KESTY N C, KUEHN M J. Incorporation of heterologous outer membrane and periplasmic proteins into Escherichia coli outer membrane vesicles[J]. J Biol Chem, 2004, 279(3): 2069-2076.
doi: 10.1074/jbc.M307628200 |
| [19] |
HAURAT M F, ADUSE-OPOKU J, RANGARAJAN M, et al. Selective sorting of cargo proteins into bacterial membrane vesicles[J]. J Biol Chem, 2011, 286(2): 1269-1276.
doi: 10.1074/jbc.M110.185744 pmid: 21056982 |
| [20] | BONNINGTON K E, KUEHN M J. Protein selection and export via outer membrane vesicles[J]. Biochim Biophys Acta, 2014, 1843(8): 1612-1619. |
| [21] |
SONG X, LV T, CHEN J, et al. Characterization of residue specific protein folding and unfolding dynamics in cells[J]. J Am Chem Soc, 2019, 141(29): 11363-11366.
doi: 10.1021/jacs.9b04435 pmid: 31305080 |
| [22] |
TAKAOKA Y, KIOI Y, MORITO A, et al. Quantitative comparison of protein dynamics in live cells and in vitro by in-cell 19F NMR[J]. Chem Comm, 2013, 49(27): 2801-2803.
doi: 10.1039/c3cc39205h |
| [23] |
WILLIAMSON M P. Using chemical shift perturbation to characterize ligand binding[J]. Prog Nucl Magn Reson Spectrosc, 2013, 73: 1-16.
doi: 10.1016/j.pnmrs.2013.02.001 |
| [1] | LIU Ying, YUAN Binhua, ZHANG Haowei. Design of a portable magnetic resonance multi-source RF pulse generator [J]. Chinese Journal of Magnetic Resonance, 0, (): 0-0. |
| [2] | ZHU Xiangwei, YANG Xue, WEI Daxiu, YAO Yefeng. In Vivo Glutathione Molecular MRS Signal Selection Based on Nuclear Spin Singlet States [J]. Chinese Journal of Magnetic Resonance, 2024, 41(4): 373-381. |
| [3] | NING Xinzhou, HUANG Zhen, CHEN Xiqu, LIU Xinjie, CHEN Gang, ZHANG Zhi, BAO Qingjia, LIU Chaoyang. Research on Transformer Super-Resolution Reconstruction Algorithm for Ultrafast Spatiotemporal Encoding Magnetic Resonance Imaging [J]. Chinese Journal of Magnetic Resonance, 2024, 41(4): 454-468. |
| [4] | PANG Qifan, WANG Zhichao, WU Yupeng, LI Jianqi. The Impact of K-Space Filling Strategy on Fat Artifacts in APT Imaging Based on FLASH Sequence [J]. Chinese Journal of Magnetic Resonance, 2024, 41(4): 443-453. |
| [5] | LUO Wenyou, RONG Xing. A W-Band Electron Paramagnetic Resonance Probe Based on Fabry-Perot Cavity [J]. Chinese Journal of Magnetic Resonance, 2024, 41(4): 393-404. |
| [6] | LI Lianjie, ZHENG Yu, XIE Shuguang, ZHANG Ming, LI Hongchuang, LIU Xiaoling, ZHAO Xiuchao, HAN Yeqing, LI Haidong, FAN Li, XIAO Yi, LIU Shiyuan, ZHOU Xin. Assessment of Pulmonary Function Changes of AECOPD with Hyperpolarized 129Xe MR [J]. Chinese Journal of Magnetic Resonance, 2024, 41(4): 363-372. |
| [7] | ZENG Xiangzheng, CHEN Junfei, HUANG Chongyang, PI Haiya, CAO Li, HUANG Zhen, GUO Wenlong, FENG Jiwen, LIU Chaoyang. Design and Research of the dDNP Automated Dissolution System [J]. Chinese Journal of Magnetic Resonance, 2024, 41(4): 382-392. |
| [8] | Hao-Yun SUN Li JiaWANG. Fusing Attention Mechanism and Dilated Convolution 3D MobileNetV2 for Classification of Hepatic Nodules [J]. Chinese Journal of Magnetic Resonance, 0, (): 0-0. |
| [9] | YANG Liming, WANG Yuanjun. Research Progress of Denoising Algorithms for Diffusion Tensor Images [J]. Chinese Journal of Magnetic Resonance, 2024, 41(3): 341-361. |
| [10] | LI Mingdao, YAO Shouquan, XU Juncheng, LV Xinglong, HE Fengcheng, JIANG Yu. Design of the Handheld NMR Console [J]. Chinese Journal of Magnetic Resonance, 2024, 41(3): 257-265. |
| [11] | WANG Xingle, SHAO Zhengze, DONG Hongchun, WEI Daxiu, CHEN Qun, YAO Yefeng. Studies on the 1H NMR Spectral Features of Hydrogen Molecules in the Interstices of SiO2 Particles [J]. Chinese Journal of Magnetic Resonance, 2024, 41(3): 315-321. |
| [12] | SHI Xing, ZHANG Yue, ZHANG Xiuli, WANG Cong. Stereochemical Research on Cembranes Diterpenoid Sinulariol Z Based on Residual Dipolar Couplings [J]. Chinese Journal of Magnetic Resonance, 2024, 41(3): 322-330. |
| [13] | GUO Xu, WANG Chenxu, ZHANG Xin, CHANG Yan, CUI Feng, GUO Qingqian, HU Tao, YANG Xiaodong. Semantic Audiovisual Single-trial Detection Based on the New Generation of Magnetoencephalography [J]. Chinese Journal of Magnetic Resonance, 2024, 41(3): 304-314. |
| [14] | LUO Qingjin, WU Liangyong, WANG Yuting, YAN Haiyang, XIANG Yifeng, CHEN Siyu. Optimization Analysis and Experimental Verification of FID NMR Coil in Polarized 3He Systems [J]. Chinese Journal of Magnetic Resonance, 2024, 41(3): 266-275. |
| [15] | YI Peng, CAO Li, HUANG Zhen, CHENG Xin, WANG Jiaxin, CHEN Li, CHEN Fang, BAO Qingjia, ZHANG Zhi, LIU Chaoyang. Development of Gradient Coils and 1H/13C Dual-resonance RF Coils for a Small-bore 5 T MRI System [J]. Chinese Journal of Magnetic Resonance, 2024, 41(3): 245-256. |
| Viewed | ||||||
|
Full text 208
|
|
|||||
|
Abstract |
|
|||||