
- Dec. 4, 2025
- Home
- About Us
- Editorial Board
- Instruction
- Subscription
- Advertisement
- Contact Us
- Chinese
- RSS

Chinese Journal of Magnetic Resonance ›› 2022, Vol. 39 ›› Issue (3): 291-302.doi: 10.11938/cjmr20212918
• Articles • Previous Articles Next Articles
Received:2021-05-13
Published:2022-09-05
Online:2021-08-27
Contact:
Yuan-jun WANG
E-mail:yjusst@126.com
CLC Number:
Qin ZHOU, Yuan-jun WANG. Groupwise Registration for Magnetic Resonance Image Based on Variational Inference[J]. Chinese Journal of Magnetic Resonance, 2022, 39(3): 291-302.
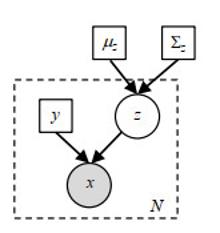
Fig.3
Probability generation model. White circle represents hidden variable, gray circle represents observed value (input image), and squares represent parameters. Dashed rectangle indicates the number of independent repeats N of the x, y, z variables inside. Registration prior is defined by the mean value μz and covariance Σz of Gaussian parameters
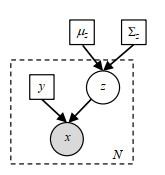
Table 1
Groupwise registration results of multiple metrics on LPBA40 test set by different algorithms
| 方法 | Avg. Dice* | GPU运行时间/s* | Percentages (%) of | EM1 | EM2 |
| ANTs (Syn) | 0.689 (0.036) | 45.6 (19) | 930.98 | 6.54 | |
| voxelmorph-diff | 0.698 (0.030) | 0.51 (0.03) | 49.6 (11) | 960.67 | 6.29 |
| voxelmorph | 0.672 (0.031) | 0.49 (0.02) | 48.9 (26) | 969.46 | 7.08 |
| 本文方法 | 0.701 (0.025) | 0.40 (0.02) | 45.0 (18) | 940.89 | 5.07 |
Fig.7
Mean images of distorted images by different algorithms. (a) Sagittal section central slice; (b) Coronal section central slice; (c) Horizontal section central slice. In each sub-figure, the first row from left to right is the mean image by voxelmorph algorithm, and voxelmorph-diff algorithm, the second row from left to right is the mean image by Syn algorithm and the method proposed in this work
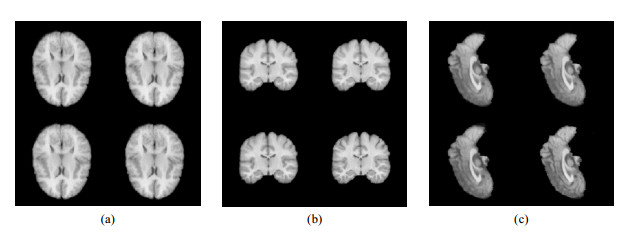
Table 2
Groupwise registration results of multiple metrics on LPBA40 testing set with different noise intensities using the proposed method
| 噪声方差(均值为0) | Avg. Dice* | GPU运行时间/s* | Percentages (%) of | EM1 | EM2 |
| 0.001 | 0.686 (0.026) | 0.50 (0.02) | 48.8 (21) | 1330.97 | 89.54 |
| 0.002 | 0.683 (0.030) | 0.60 (0.04) | 49.3 (27) | 1360.67 | 100.29 |
| 0.003 | 0.646 (0.031) | 0.58 (0.02) | 51.7 (19) | 1469.46 | 99.08 |
| 0.004 | 0.656 (0.020) | 0.62 (0.03) | 54.5 (28) | 1480.89 | 113.07 |
| 1 |
WANG Y, JIANG F, LIU YReference-free brain template construction with population symmetric registration[J]. Med Biol Eng Comput, 2020,58 (9): 2083- 2093.
doi: 10.1007/s11517-020-02226-5 |
| 2 |
MARTÍN-GONZÁLEZ E, SEVILLA T, REVILLA-ORODEA A, et alGroupwise non-rigid registration with deep learning: an affordable solution applied to 2D cardiac cine MRI reconstruction[J]. Entropy, 2020,22 (6): 687.
doi: 10.3390/e22060687 |
| 3 |
GEE J C, REIVICH M, BILANIUK L, et alEvaluation of multiresolution elastic matching using mri data[J]. Proc Spie, 1991,1445,226- 234.
doi: 10.1117/12.45220 |
| 4 |
RUECKERT D, SONODA L INonrigid registration using free-form deformations: application to breast mr images[J]. IEEE T Med Imaging, 1999,18 (8): 712- 721.
doi: 10.1109/42.796284 |
| 5 | THIRION J PImage matching as a diffusion process: an analogy with Maxwell's demons[J]. Med Image Anal, 2011,2 (3): 243- 260. |
| 6 |
ZHONG J, PHUA D, QIU AQuantitative evaluation of lddmm, freesurfer, and caret for cortical surface mapping[J]. Neuroimage, 2010,52 (1): 131- 141.
doi: 10.1016/j.neuroimage.2010.03.085 |
| 7 |
ASAMI T, BOUIX S, WHITFORD T J, et alLongitudinal loss of gray matter volume in patients with first-episode schizophrenia: dartel automated analysis and roi validation[J]. Neuroimage, 2012,59 (2): 986- 996.
doi: 10.1016/j.neuroimage.2011.08.066 |
| 8 |
GUIMOND A, MEUNIER J, THIRION J PAverage brain models: a convergence study[J]. Comput Vis Image Und, 2000,77 (2): 192- 210.
doi: 10.1006/cviu.1999.0815 |
| 9 | SEGHERS D, D'AGOSTINO E, MAES F, et al. Construction of a brain template from mr images using state-of-the-art registration and segmentation techniques[C]// Medical Image Computing and Computer-Assisted Intervention--MICCAI 2004, 7th International Conference Saint-Malo, France, September 26-29, 2004, Proceedings, Part I. 2004. |
| 10 |
WU G, JIA H, WANG Q, et alSharpmean: groupwise registration guided by sharp mean image and tree-based registration[J]. Neuroimage, 2011,56 (4): 1968- 1981.
doi: 10.1016/j.neuroimage.2011.03.050 |
| 11 |
WU G, WANG Q, JIA H, et alFeature-based groupwise registration by hierarchical anatomical correspondence detection[J]. Hum Brain Mapp, 2012,33 (2): 253- 271.
doi: 10.1002/hbm.21209 |
| 12 | YANOVSKY I, THOMPSON P M, OSHER S, et al. Topology preserving log-unbiased nonlinear image registration: theory and implementation[C]// IEEE Conference on Computer Vision & Pattern Recognition. Minneapolis, Minnesota, USA: IEEE, 2007. |
| 13 | WANG Q, CHEN L Y YAP P T, et alGroupwise registration based on hierarchical image clustering and atlas synthesis[J]. Hum Brain Mapp, 2010,31,1128- 1140. |
| 14 |
CHE T, ZHENG Y, CONG J, et alDeep group-wise registration for multi-spectural images from fundus images[J]. IEEE Access, 2019,7,27650- 27661.
doi: 10.1109/ACCESS.2019.2901580 |
| 15 | CHE T, ZHENG Y, SUI X, et al. Dgr-net: deep groupwise registration of multispectral images[C]// Information Processing in Medical Imaging - 26th International Conference , Hong Kong, china: IPMI, 2019: 706-717. |
| 16 | HAASE R, HELDMANN S, LELLMANN J. Deformable groupwise image registration using low-rank and sparse decomposition[EB/OL]. [2020-06-10].https://arxiv.org/abs/2001.03509. |
| 17 | DALCA A V, RAKIC M, GUTTAG J, et al. Learning conditional deformable templates with convolutional networks[C]//Neural Information Processing Systems 2019(NeurIPS 2019), Vancouver, BC, Canada, 2019: 804-816. |
| 18 | YU E M, DALCA A V, SABUNCU M R Learning conditional deformable shape templates for brain anatomy[M]. Lima: Machine Learning in Medical Imaging, 2020, 353- 352. |
| 19 | SIEBERT H, HEINRICH M P Deep groupwise registration of mri using deforming autoencoders[M]. Berlin: Springer, 2020, 236- 241. |
| 20 | HE Z Y, CHUNG A C S. Unsupervised end-to-end groupwise registration framework without generating templates[C]// 2020 IEEE International Conference on Image Processing (ICIP). IEEE, 2020: 375-379. |
| 21 |
DALCA A V, BALAKRISHNAN G, GUTTAG J, et alUnsupervised learning of probabilistic diffeomorphic registration for images and surfaces[J]. Med Image Anal, 2019,57,226- 236.
doi: 10.1016/j.media.2019.07.006 |
| 22 | BALAKRISHNAN G, ZHAO A, SABUNCU M R, et alVoxelmorph: a learning framework for deformable medical image registration[J]. IEEE T Med Imaging, 2019,1788- 1800. |
| 23 |
SHATTUCK D W, MIRZA M, ADISETIYO V, et alConstruction of a 3d probabilistic atlas of human cortical structures.[J]. Neuroimage, 2008,39 (3): 1064- 1080.
doi: 10.1016/j.neuroimage.2007.09.031 |
| 24 | 王远军, 刘玉基于图像集拓扑中心的群体配准方法[J]. 波谱学杂志, 2018,35 (4): 60- 67. |
| WANG Y J, LIU YGroup registration method based on topological center of images[J]. Chinese J Magn Reson, 2018,35 (4): 60- 67. | |
| 25 | 蔡文琴, 王远军基于磁共振成像的人脑图谱构建方法研究进展[J]. 波谱学杂志, 2020,37 (2): 241- 253. |
| CAI W Q, WANG Y JAdvances in the construction of the human brain map based on magnetic resonance imaging[J]. Chinese J Magn Reson, 2020,37 (2): 241- 253. | |
| 26 | 刘可文, 刘紫龙, 汪香玉, 等基于级联卷积神经网络的前列腺磁共振图像分类[J]. 波谱学杂志, 2020,37 (2): 152- 161. |
| LIU K W, LIU Z L, WANG X Y, et alProstate magnetic resonance image classification based on cascading convolutional neural networks[J]. Chinese J Magn Reso, 2020,37 (2): 152- 161. |
| [1] | CAO Fei, XU Qianqian, CHEN Hao, ZU Jie, LI Xiaowen, TIAN Jin, BAO Lei. An Intelligent Diagnosis Method for NIID Based on Cross Self-supervision and DWI [J]. Chinese Journal of Magnetic Resonance, 2025, 42(2): 154-163. |
| [2] | XUE Peiyang, GENG Chen, LI Yuxin, BAO Yifang, LU Yucheng, DAI Yakang. A Classification Method for Cerebral Aneurysms in TOF-MRA Based on Improved 3D ResNet50 Model [J]. Chinese Journal of Magnetic Resonance, 2025, 42(1): 56-66. |
| [3] | NING Xinzhou, HUANG Zhen, CHEN Xiqu, LIU Xinjie, CHEN Gang, ZHANG Zhi, BAO Qingjia, LIU Chaoyang. Research on Transformer Super-Resolution Reconstruction Algorithm for Ultrafast Spatiotemporal Encoding Magnetic Resonance Imaging [J]. Chinese Journal of Magnetic Resonance, 2024, 41(4): 454-468. |
| [4] | YANG Liming, WANG Yuanjun. Research Progress of Denoising Algorithms for Diffusion Tensor Images [J]. Chinese Journal of Magnetic Resonance, 2024, 41(3): 341-361. |
| [5] | Dai Junlong, He Cong, Wu Jie, Bian Yun. Pancreatic Cystic Neoplasms Segmentation Network Combining Dual Decoding and Global Attention Upsampling Modules [J]. Chinese Journal of Magnetic Resonance, 2024, 41(2): 151-161. |
| [6] | YANG Yu, CHEN Bo, WU Liubin, LIN Enping, HUANG Yuqing, CHEN Zhong. Spectrum Reconstruction for Laplace NMR: From Handcraft Regularization to Deep Learning [J]. Chinese Journal of Magnetic Resonance, 2024, 41(2): 191-208. |
| [7] | CHANG Bo, SUN Haoyun, GAO Qingyu, WANG Lijia. Research Progress on Cardiac Segmentation in Different Modal Medical Images by Traditional Methods and Deep Learning [J]. Chinese Journal of Magnetic Resonance, 2024, 41(2): 224-244. |
| [8] | XU Zhenshun, YUAN Xiaohan, HUANG Ziheng, SHAO Chengwei, WU Jie, BIAN Yun. Multi-source Feature Classification Model of Pancreatic Mucinous and Serous Cystic Neoplasms Based on Deep Learning [J]. Chinese Journal of Magnetic Resonance, 2024, 41(1): 19-29. |
| [9] | LAI Jiawen, WANG Yuling, CAI Xiaoyu, ZHOU Lihua. Multidimensional Information Fusion Method for Meniscal Tear Classification Based on CNN-SVM [J]. Chinese Journal of Magnetic Resonance, 2023, 40(4): 423-434. |
| [10] | WANG Hui, WANG Tiantian, WANG Lijia. Squeeze-and-excitation Residual U-shaped Network for Left Myocardium Segmentation Based on Cine Cardiac Magnetic Resonance Images [J]. Chinese Journal of Magnetic Resonance, 2023, 40(4): 435-447. |
| [11] | Li Yijie, YANG Xinyu, YANG Xiaomei. Magnetic Resonance Image Reconstruction of Multi-scale Residual Unet Fused with Attention Mechanism [J]. Chinese Journal of Magnetic Resonance, 2023, 40(3): 307-319. |
| [12] | LU Qiqi, LIAN Zifeng, LI Jialong, SI Wenbin, MAI Zhaohua, FENG Yanqiu. Magnetic Resonance R2* Parameter Mapping of Liver Based on Self-supervised Deep Neural Network [J]. Chinese Journal of Magnetic Resonance, 2023, 40(3): 258-269. |
| [13] | ZHANG Jiajun, LU Yucheng, BAO Yifang, LI Yuxin, GENG Chen, HU Fuyuan, DAI Yakang. An Automatic Segmentation Method of Cerebral Arterial Tree in TOF-MRA Based on DBCNet [J]. Chinese Journal of Magnetic Resonance, 2023, 40(3): 320-331. |
| [14] | TIAN Hui, WU Jie, BIAN Yun, ZHANG Zhiwei, SHAO Chengwei. Classification of Pancreatic Cystic Tumors Based on DenseNet and Transfer Learning [J]. Chinese Journal of Magnetic Resonance, 2023, 40(3): 270-279. |
| [15] | HUANG Min,LI Siyi,CHEN Junbo,ZHOU Dao. Progress of Magnetic Resonance Fingerprinting Technology and Its Clinical Application [J]. Chinese Journal of Magnetic Resonance, 2023, 40(2): 207-219. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
