
- Jul. 10, 2025
- Home
- About Us
- Editorial Board
- Instruction
- Subscription
- Advertisement
- Contact Us
- Chinese
- RSS

Chinese Journal of Magnetic Resonance ›› 2022, Vol. 39 ›› Issue (4): 381-392.doi: 10.11938/cjmr20222974
• Articles • Previous Articles Next Articles
Yun-shan PEI1,2,Cai ZHANG1,2,Xiao-li LIU1,Kai CHENG1,Ze-ting ZHANG1,*( ),Cong-gang LI1
),Cong-gang LI1
Received:2022-01-28
Online:2022-12-05
Published:2022-03-10
Contact:
Ze-ting ZHANG
E-mail:zhangzeting@wipm.ac.cn
CLC Number:
Yun-shan PEI, Cai ZHANG, Xiao-li LIU, Kai CHENG, Ze-ting ZHANG, Cong-gang LI. Inhibition of α-Synuclein Aggregation by the Interaction Between Protein Disulfide Isomerase and α-Synuclein[J]. Chinese Journal of Magnetic Resonance, 2022, 39(4): 381-392.

Fig.1
The b'xa' domain of PDI interacts with N-terminal domain of αsyn. (a) Model for PDI (red) primary sequence, PDI ab domain (pink) and PDI b'xa' domain (blue) are shown underneath. (b) Surface crystal structure of human PDI (PDB ID: 4EKZ). (c) Protein purities analyzed by SDS-PAGE. M, Marker; lane 1, αsyn1-60; lane 2, αsyn; lane 3, PDI ab; lane 4, PDI b'xa'; lane 5, PDI. (d) and (e) Spectral overlay of αsyn in the absence (black) and presence of equivalent amount of PDI (red) and PDI b'xa' (blue). Residues showing significant signal attenuation or shifts were marked. (f) Residue-resolved attenuation (I/I0) of αsyn in the presence of PDI (red), PDI ab (pink) and PDI b'xa' (blue). (g) Hydrophobic residues of αsyn predicted by OMH method, the binding and hydrophobic areas are colored gray
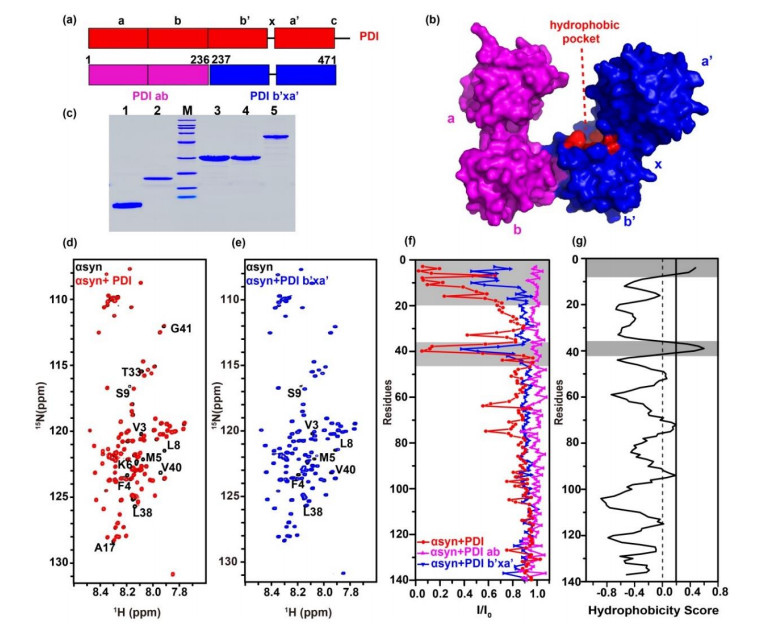

Fig.2
The effect on αsyn aggregation of PDI, PDI ab and PDI b'xa' detected by ThT experiment. (a) Aggregations kinetics of αsyn in the absence (black) and presence of equimolar PDI ab (pink), PDI b'xa' (blue) and PDI (red). (b) The fitted half-time (t1/2) values of aggregation and their statistical analysis. ns: not significant, ****: p < 0.0001; the dots represent the values obtained from four parallel experiments
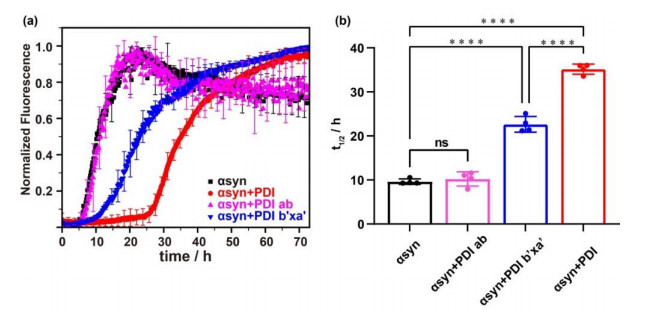
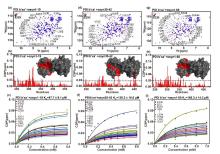
Fig.3
NMR results showing the interaction between PDI b'xa' with the N-terminal domain of αsyn. Overlaid 1H-15N TROSY spectra of PDI b'xa' (red) with (blue) αsyn 1-19 (a), αsyn 30-42 (d) or αsyn 1-60 (g). E239 were selected as representative residues for global chemical shift perturbations (CSPs) during NMR titration. Residue-resolved CSPs of PDI b'xa' during titrations with αsyn 1-19(b), 30-42(e) and 1-60 (h). KD values were fitted from curves of residues with significant CSPs, as a function of αsyn 1-19(c), αsyn 30-42(f), and αsyn 1-60 (i) concentration. Residues showing significant CSPs were marked in spectra and mapped red on the crystal structure surface of PDI b'xa'. Threshold level was indicated as 'mean+standard deviation'. Disperse residues were verified and removed by the threshold of 'mean +2×standard deviation'
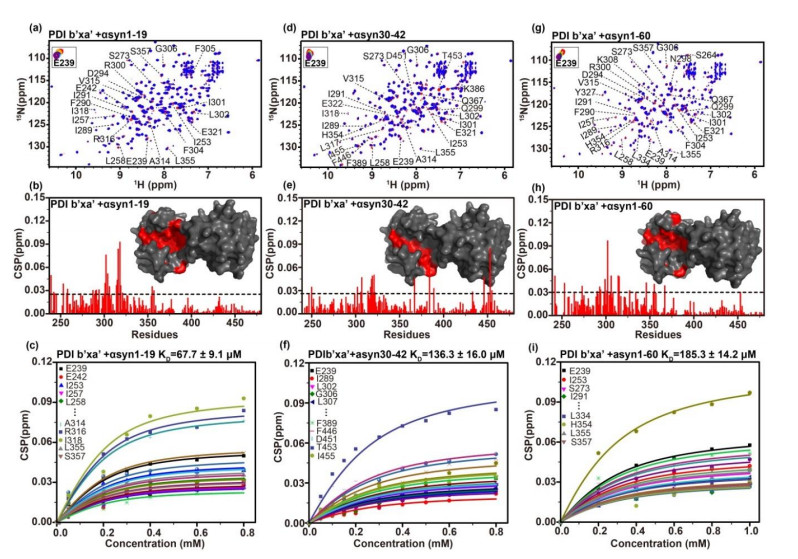
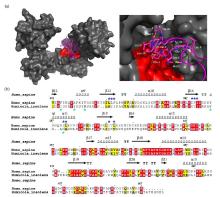
Fig.4
Hydrophobic binding mode of PDI with the N-terminal domain of αsyn. (a) Docking model of PDI binding αsyn1-40 (left panel) and a local zoom view of binding area (right panel). The relevant residues of αsyn1-40 involving in binding PDI were depicted as magenta sticks. HDOCK experiments were performed on the web server (http://hdock.phys.hust.edu.cn/) by uploading human PDI crystal structure, amino acid sequences of αsyn1-40 and their binding sites identified by NMR titration. Optimal docking results were plotted with PyMol software, PDI molecular surface was colored gray and its hydrophobic pocket was marked in red, the magenta cartoon represented αsyn1-40 structure. (b) Alignment of amino acid sequence of human PDI b'xa' from HiPDI b'xa'. Sequence identity and similarity were calculated by Clustalw[52] and ESPript 3.0[53]. Similar amino acids were showed as black letters in yellow boxes and identical amino acids were showed as white letters in red boxes. The secondary structural domains were indicated on the top of the sequences. Blue and green asterisks represented the key hydrophobic residues of αsyn interacting with human PDI and HiPDI, respectively

| 1 |
THEILLET F X, BINOLFI A, BEKEI B, et al Structural disorder of monomeric α-synuclein persists in mammalian cells[J]. Nature, 2016, 530 (7588): 45- 50.
doi: 10.1038/nature16531 |
| 2 |
WEINREB P H, ZHEN W, POON A W, et al NACP, a protein implicated in Alzheimer's disease and learning, is natively unfolded[J]. Biochemistry, 1996, 35 (43): 13709- 13715.
doi: 10.1021/bi961799n |
| 3 |
IADANZA M G, JACKSON M P, HEWITT E W, et al A new era for understanding amyloid structures and disease[J]. Nat Rev Mol Cell Biol, 2018, 19 (12): 755- 773.
doi: 10.1038/s41580-018-0060-8 |
| 4 |
LIU C W, GIASSON B I, LEWIS K A, et al A precipitating role for truncated alpha-synuclein and the proteasome in alpha-synuclein aggregation - Implications for pathogenesis of Parkinson disease[J]. J Biol Chem, 2005, 280 (24): 22670- 22678.
doi: 10.1074/jbc.M501508200 |
| 5 |
DETTMER U, SELKOE D, BARTELS T New insights into cellular α-synuclein homeostasis in health and disease[J]. Curr Opin Neurobiol, 2016, 36, 15- 22.
doi: 10.1016/j.conb.2015.07.007 |
| 6 |
SPILLANTINI M G, SCHMIDT M L, LEE V M Y, et al α-Synuclein in Lewy bodies[J]. Nature, 1997, 388 (6645): 839- 840.
doi: 10.1038/42166 |
| 7 |
LAUTENSCHL GER J, KAMINSKI C F, KAMINSKI SCHIERLE G S α-synuclein – regulator of exocytosis, endocytosis, or both?[J]. Trends in Cell Biol, 2017, 27 (7): 468- 479.
doi: 10.1016/j.tcb.2017.02.002 |
| 8 |
MAHUL-MELLIER A L, BURTSCHER J, MAHARJAN N, et al The process of Lewy body formation, rather than simply α-synuclein fibrillization, is one of the major drivers of neurodegeneration[J]. Proc Natl Acad Sci U S A, 2020, 117 (9): 4971- 4982.
doi: 10.1073/pnas.1913904117 |
| 9 |
SHAHMORADIAN S H, LEWIS A J, GENOUD C, et al Lewy pathology in Parkinson's disease consists of crowded organelles and lipid membranes[J]. Nat Neurosci, 2019, 22 (7): 1099- 1109.
doi: 10.1038/s41593-019-0423-2 |
| 10 |
BARTELS T, AHLSTROM L S, LEFTIN A, et al The N-terminus of the intrinsically disordered protein α-synuclein triggers membrane binding and helix folding[J]. Biophys J, 2010, 99 (7): 2116- 2124.
doi: 10.1016/j.bpj.2010.06.035 |
| 11 |
BURMANN B M, GEREZ J A, MATEČKO-BURMANN I, et al Regulation of α-synuclein by chaperones in mammalian cells[J]. Nature, 2020, 577 (7788): 127- 132.
doi: 10.1038/s41586-019-1808-9 |
| 12 |
GRUSCHUS J M, YAP T L, PISTOLESI S, et al NMR structure of calmodulin complexed to an N-terminally acetylated α-synuclein peptide[J]. Biochemistry, 2013, 52 (20): 3436- 3445.
doi: 10.1021/bi400199p |
| 13 |
GIASSON B I, MURRAY I V, TROJANOWSKI J Q, et al A hydrophobic stretch of 12 amino acid residues in the middle of alpha-synuclein is essential for filament assembly[J]. J Biol Chem, 2001, 276 (4): 2380- 2386.
doi: 10.1074/jbc.M008919200 |
| 14 |
BINOLFI A, RASIA R M, BERTONCINI C W, et al Interaction of alpha-synuclein with divalent metal ions reveals key differences: A link between structure, binding specificity and fibrillation enhancement[J]. J Am Chem Soc, 2006, 128 (30): 9893- 9901.
doi: 10.1021/ja0618649 |
| 15 |
NIELSEN M S, VORUM H, LINDERSSON E, et al Ca2+ binding to alpha-synuclein regulates ligand binding and oligomerization[J]. J Biol Chem, 2001, 276 (25): 22680- 22684.
doi: 10.1074/jbc.M101181200 |
| 16 |
BINOLFI A, RASIA R M, BERTONCINI C W, et al Interaction of alpha-synuclein with divalent metal ions reveals key differences: a link between structure, binding specificity and fibrillation enhancement[J]. J Am Chem Soc, 2006, 128 (30): 9893- 9901.
doi: 10.1021/ja0618649 |
| 17 |
SERRANO A, QIAO X, MATOS J O, et al Reversal of alpha-synuclein fibrillization by protein disulfide isomerase[J]. Front Cell Dev Biol, 2020, 8, 726.
doi: 10.3389/fcell.2020.00726 |
| 18 |
DEDMON M M, CHRISTODOULOU J, WILSON M R, et al Heat shock protein 70 inhibits alpha-synuclein fibril formation via preferential binding to prefibrillar species[J]. J Biol Chem, 2005, 280 (15): 14733- 14740.
doi: 10.1074/jbc.M413024200 |
| 19 |
DIMANT H, EBRAHIMI-FAKHARI D, MCLEAN P J Molecular chaperones and co-chaperones in Parkinson disease[J]. Neuroscientist, 2012, 18 (6): 589- 601.
doi: 10.1177/1073858412441372 |
| 20 |
GAO X, CARRONI M, NUSSBAUM-KRAMMER C, et al Human Hsp70 disaggregase reverses Parkinson's-linked α-synuclein amyloid fibrils[J]. Mol Cell, 2015, 59 (5): 781- 793.
doi: 10.1016/j.molcel.2015.07.012 |
| 21 |
PEMBERTON S, MADIONA K, PIERI L, et al Hsc70 protein interaction with soluble and fibrillar alpha-synuclein[J]. J Biol Chem, 2011, 286 (40): 34690- 34699.
doi: 10.1074/jbc.M111.261321 |
| 22 |
RANJAN P, KUMAR A The involvement of His50 during protein disulfide isomerase binding is essential for inhibiting α-Syn fibril formation[J]. Biochemistry, 2016, 55 (19): 2677- 2680.
doi: 10.1021/acs.biochem.6b00280 |
| 23 |
UEHARA T, NAKAMURA T, YAO D, et al S-nitrosylated protein-disulphide isomerase links protein misfolding to neurodegeneration[J]. Nature, 2006, 441 (7092): 513- 517.
doi: 10.1038/nature04782 |
| 24 |
WANG C, LI W, REN J, et al Structural insights into the redox-regulated dynamic conformations of human protein disulfide isomerase[J]. Antioxid Redox Signal, 2013, 19 (1): 36- 45.
doi: 10.1089/ars.2012.4630 |
| 25 |
DARBY N J, CREIGHTON T E Functional properties of the individual thioredoxin-like domains of protein disulfide isomerase[J]. Biochemistry, 1995, 34 (37): 11725- 11735.
doi: 10.1021/bi00037a009 |
| 26 |
KLAPPA P, RUDDOCK L W, DARBY N J, et al The b' domain provides the principal peptide-binding site of protein disulfide isomerase but all domains contribute to binding of misfolded proteins[J]. EMBO J, 1998, 17 (4): 927- 935.
doi: 10.1093/emboj/17.4.927 |
| 27 |
YAO Y, ZHOU Y, WANG C Both the isomerase and chaperone activities of protein disulfide isomerase are required for the reactivation of reduced and denatured acidic phospholipase A2[J]. EMBO J, 1997, 16 (3): 651- 658.
doi: 10.1093/emboj/16.3.651 |
| 28 |
ELLGAARD L, RUDDOCK L W The human protein disulphide isomerase family: substrate interactions and functional properties[J]. EMBO Rep, 2005, 6 (1): 28- 32.
doi: 10.1038/sj.embor.7400311 |
| 29 |
PAPP E, NARDAI G, STI C, et al Molecular chaperones, stress proteins and redox homeostasis[J]. Biofactors, 2003, 17 (1-4): 249- 257.
doi: 10.1002/biof.5520170124 |
| 30 |
CHEN X, ZHANG X, LI C, et al S-nitrosylated protein disulfide isomerase contributes to mutant SOD1 aggregates in amyotrophic lateral sclerosis[J]. J Neurochem, 2013, 124 (1): 45- 58.
doi: 10.1111/jnc.12046 |
| 31 |
CHENG H, WANG L, WANG C C Domain a' of protein disulfide isomerase plays key role in inhibiting alpha-synuclein fibril formation[J]. Cell Stress Chaperones, 2010, 15 (4): 415- 421.
doi: 10.1007/s12192-009-0157-2 |
| 32 |
YAGI-UTSUMI M, SATOH T, KATO K Structural basis of redox-dependent substrate binding of protein disulfide isomerase[J]. Sci Rep, 2015, 5, 13909.
doi: 10.1038/srep13909 |
| 33 | 余锦波, 张偲, 张则婷, 等 Alpha-突触核蛋白与完整线粒体相互作用的NMR研究[J]. 波谱学杂志, 2021, 38 (2): 164- 172. |
| YU J B, ZHANG C, ZHANG Z T, et al Interactions between α-synuclein and intact mitochondria studied by NMR[J]. Chinese J Magn Reson, 2021, 38 (2): 164- 172. | |
| 34 | 寇新慧, 刘乙祥, 刘兴弘, 等 探测应答调控蛋白PhoBNF20D自由态中存在的Pre-Active构象[J]. 波谱学杂志, 2019, 36 (2): 164- 171. |
| KOU X H, LIU Y X, LIU X H, et al Visualizing the pre-active conformation of response regulator PhoBNF20D in its apo state[J]. Chinese J Magn Reson, 2019, 36 (2): 164- 171. | |
| 35 |
JAO C C, DER-SARKISSIAN A, CHEN J, et al Structure of membrane-bound alpha-synuclein studied by site-directed spin labeling[J]. Proc Natl Acad Sci U S A, 2004, 101 (22): 8331- 8336.
doi: 10.1073/pnas.0400553101 |
| 36 |
HOYER W, ANTONY T, CHERNY D, et al Dependence of alpha-synuclein aggregate morphology on solution conditions[J]. J Mol Biol, 2002, 322 (2): 383- 393.
doi: 10.1016/S0022-2836(02)00775-1 |
| 37 | 戴晨晔, 刘买利, 李从刚 低盐和高盐环境下α-synuclein构象的19F NMR研究[J]. 波谱学杂志, 2015, 32 (1): 33- 44. |
| DAI C Y, LIU M L, LI C G Salt content-dependent conformational changes of alpha-synuclein studied by 19F NMR[J]. Chinese J Magn Reson, 2015, 32 (1): 33- 44. | |
| 38 |
PEI Y, LIU X, CHENG K, et al Backbone resonance assignment of PDI b'xa' domain construct[J]. Biomol NMR Assign, 2021, 15 (2): 409- 413.
doi: 10.1007/s12104-021-10038-3 |
| 39 | DELAGLIO F, GRZESIEK S, VUISTER G W, et al NMRPipe: A multidimensional spectral processing system based on UNIX pipes[J]. J Biomol NMR, 1995, 6 (3): 277- 293. |
| 40 |
LEE W, TONELLI M, MARKLEY J L NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy[J]. Bioinformatics, 2015, 31 (8): 1325- 1327.
doi: 10.1093/bioinformatics/btu830 |
| 41 |
WILLIAMSON, MIKE P Using chemical shift perturbation to characterise ligand binding[J]. Prog Nucl Magn Reson Spectrosc, 2013, 73, 1- 16.
doi: 10.1016/j.pnmrs.2013.02.001 |
| 42 |
ARAI M, FERREON J C, WRIGHT P E Quantitative analysis of multisite protein-ligand interactions by NMR: binding of intrinsically disordered p53 transactivation subdomains with the TAZ2 domain of CBP[J]. J Am Chem Soc, 2012, 134 (8): 3792- 3803.
doi: 10.1021/ja209936u |
| 43 |
DE OLIVEIRA G A P, SILVA J L Alpha-synuclein stepwise aggregation reveals features of an early onset mutation in Parkinson's disease[J]. Commun Biol, 2019, 2, 374.
doi: 10.1038/s42003-019-0598-9 |
| 44 |
CAMPIONI S, CARRET G, JORDENS S, et al The presence of an air-water interface affects formation and elongation of α-synuclein fibrils[J]. J Am Chem Soc, 2014, 136 (7): 2866- 2875.
doi: 10.1021/ja412105t |
| 45 |
CROKE R, PATIL S, QUEVREAUX J, et al NMR determination of pK(a) values in α-synuclein[J]. Protein sci, 2011, 20 (2): 256- 269.
doi: 10.1002/pro.556 |
| 46 |
SWEET R M, EISENBERG D Correlation of sequence hydrophobicities measures similarity in three-dimensional protein structure[J]. J Mol Biol, 1983, 171 (4): 479- 488.
doi: 10.1016/0022-2836(83)90041-4 |
| 47 |
BIANCALANA M, KOIDE S Molecular mechanism of thioflavin-T binding to amyloid fibrils[J]. Biochim Biophys Acta, 2010, 1804 (7): 1405- 1412.
doi: 10.1016/j.bbapap.2010.04.001 |
| 48 |
KHURANA R, COLEMAN C, IONESCU-ZANETTI C, et al Mechanism of thioflavin T binding to amyloid fibrils[J]. J Struct Biol, 2005, 151 (3): 229- 238.
doi: 10.1016/j.jsb.2005.06.006 |
| 49 | 陈艳华, 张则婷, 白佳, 等 PDI抑制α-synuclein纤维化聚集作用机制研究[J]. 波谱学杂志, 2017, 34 (2): 131- 136. |
| CHEN Y H, ZHANG Z T, BAI J, et al Inhibition mechanisms of protein disulfide isomerase on α-synuclein fibril aggregation[J]. Chinese J Magn Reson, 2017, 34 (2): 131- 136. | |
| 50 |
BYRNE LEE J, SIDHU A, WALLIS A K, et al Mapping of the ligand-binding site on the b′ domain of human PDI: interaction with peptide ligands and the x-linker region[J]. Biochem J, 2009, 423 (2): 209- 217.
doi: 10.1042/BJ20090565 |
| 51 |
YAN Y, ZHANG D, ZHOU P, et al HDOCK: a web server for protein-protein and protein-DNA/RNA docking based on a hybrid strategy[J]. Nucleic Acids Res, 2017, 45 (W1): W365- W373.
doi: 10.1093/nar/gkx407 |
| 52 |
LARKIN M A, BLACKSHIELDS G, BROWN NP, et al Clustal W and Clustal X version 2.0[J]. Bioinformatics, 2007, 23 (21): 2947- 2948.
doi: 10.1093/bioinformatics/btm404 |
| 53 |
ROBERT X, GOUET P Deciphering key features in protein structures with the new ENDscript server[J]. Nucleic Acids Res, 2014, 42 (W1): W320- W324.
doi: 10.1093/nar/gku316 |
| 54 |
DOHERTY C P A, ULAMEC S M, MAYA-MARTINEZ R, et al A short motif in the N-terminal region of α-synuclein is critical for both aggregation and function[J]. Nat Struct Mol Biol, 2020, 27 (3): 249- 259.
doi: 10.1038/s41594-020-0384-x |
| [1] | Xiao-yang ZHANG, Shou-quan YAO, Jun-cheng XU, Yu JIANG. Magnetic Field Locking System Based on Fluxgate and Time Domain Digital Frequency Discrimination [J]. Chinese Journal of Magnetic Resonance, 2022, 39(4): 448-458. |
| [2] | Han HU,Wei-yu WANG,Jun XU,Feng DENG. 1, 3-Butadienen Hydrogenation on Supported Pd-Sn Bimetallic Catalysts Investigated by Parahydrogen-induced Polarization [J]. Chinese Journal of Magnetic Resonance, 2022, 39(2): 133-143. |
| [3] | Qian XU,Lang CHEN,Xiang-ying HU,Cong-gang LI,Yi-xiang LIU,Ling JIANG. The Effect of T69E-mimicked Phosphorylation on the Interaction Between Bcl-2 and Nur77 [J]. Chinese Journal of Magnetic Resonance, 2022, 39(1): 87-95. |
| [4] | Xiao-qing LIN,Shi-jia DU,Hao-lin ZHAN,Yu-qing HUANG,Zhong CHEN. Two-Dimensional Homonuclear Orthogonal-Pattern Phase-Sensitive J-Resolved NMR Spectroscopy Based on Pure Shifts [J]. Chinese Journal of Magnetic Resonance, 2021, 38(4): 448-459. |
| [5] | Yao XIAO,Chang-jiu XIA,Xian-feng YI,Feng-qing LIU,Shang-bin LIU,An-min ZHENG. Progress in the Studies on Sn-Zeolites by Solid-State Nuclear Magnetic Resonance [J]. Chinese Journal of Magnetic Resonance, 2021, 38(4): 571-584. |
| [6] | Xiao-dong HU,Wen-xian LAN,Chun-xi WANG,Chun-yang CAO. Research Advance and NMR Studies of Anti-Cancer Small Molecules Targeting c-MYC G4-DNA [J]. Chinese Journal of Magnetic Resonance, 2021, 38(4): 503-513. |
| [7] | Jia-min WU,Yu-cheng HE,Zheng XU,Yan-he ZHU,Wen-zheng JIANG. A Wide-Band Matching Method for Radio Frequency Coils Used in Soil Moisture Measurement [J]. Chinese Journal of Magnetic Resonance, 2021, 38(3): 414-423. |
| [8] | Zi-hao WANG,He XU,Tao WANG,Shan-zhong YANG,Yun-sheng DING,Hai-bing WEI. NMR Spectroscopic Studies on (exo, endo) C-2 Monosubstituted Norbornene Derivatives [J]. Chinese Journal of Magnetic Resonance, 2021, 38(3): 323-335. |
| [9] | Chong-wu WANG,Xi HUANG,Lei SHI,Shi-zhen CHEN,Xin ZHOU. Cathepsin B Triggered Hyperpolarization 129Xe MRI Probe for Ultra-Sensitive Lung Cancer Cells Detection [J]. Chinese Journal of Magnetic Resonance, 2021, 38(3): 336-344. |
| [10] | Yi LI,Jia-xiang XIN,Jia-chen WANG,Da-xiu WEI,Ye-feng YAO. Preparation Efficiency of Nuclear Spin Singlet State: A Comparison Among Three Pulse Sequences [J]. Chinese Journal of Magnetic Resonance, 2021, 38(2): 227-238. |
| [11] | Jin-bo YU,Cai ZHANG,Ze-ting ZHANG,Guo-hua XU,Cong-gang LI. Interactions Between α-synuclein and Intact Mitochondria Studied by NMR [J]. Chinese Journal of Magnetic Resonance, 2021, 38(2): 164-172. |
| [12] | Wei ZHANG,Yi-ming WU,Wei-ping CUI,Liang XIAO. Correction for the Nuclear Magnetic Resonance Porosity in Heavy Oil-bearing Reservoirs [J]. Chinese Journal of Magnetic Resonance, 2021, 38(2): 204-214. |
| [13] | Kun MENG,Sheng-jian WANG,Zong-an XUE,Rui-qing HOU,Liang XIAO. Quantitative Evaluation of Shale Pore Structure Using Nuclear Magnetic Resonance Data [J]. Chinese Journal of Magnetic Resonance, 2021, 38(2): 215-226. |
| [14] | Zhi-wu ZHANG,Ju YANG,Ze-feng NIE,Shang-xiang YE,Xu DONG,Chun TANG. Development of a Temperature Senor Based on 19F-labeled Phosphorylated Ubiquitin [J]. Chinese Journal of Magnetic Resonance, 2021, 38(2): 173-181. |
| [15] | Xin-yi ZHAO,Dong HAN,Hong-jun LUO,Wen-bin SHEN,Gong-jun YANG. Spectroscopic Studies of Delafloxacin Meglumine [J]. Chinese Journal of Magnetic Resonance, 2021, 38(2): 268-276. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||