
- Aug. 14, 2025
- Home
- About Us
- Editorial Board
- Instruction
- Subscription
- Advertisement
- Contact Us
- Chinese
- RSS

Chinese Journal of Magnetic Resonance ›› 2023, Vol. 40 ›› Issue (3): 246-257.doi: 10.11938/cjmr20222996
• Articles • Previous Articles Next Articles
ZHAO Beibei1,2,ZHAN Jianhua1,2,HU Qin1,2,ZHU Qinjun1,LIU Maili1,2,3,4,ZHANG Xu1,2,3,4,*( )
)
Received:2022-04-10
Published:2023-09-05
Online:2022-05-12
Contact:
*Tel: 027-87197056, E-mail: CLC Number:
ZHAO Beibei, ZHAN Jianhua, HU Qin, ZHU Qinjun, LIU Maili, ZHANG Xu. NMR Study on the Mechanism of Cytochrome c Methionine Oxidation[J]. Chinese Journal of Magnetic Resonance, 2023, 40(3): 246-257.

Fig. 2
1H-13C HSQC spectra of 13C full-labeled and methyl-13C methionine labeled cytochrome c under different states. (a) 0.1 mmol/L 13C full-labeled cytochrome c, oxidative state; (b) 0.1 mmol/L methyl- 13C labeling methione labeled cytochrome c, oxidative state; (c) 0.1 mmol/L 13C full-labeled cytochrome c, reductive state; (d) 0.1 mmol/L methyl- 13C methionine labeled cytochrome c, reductive state. (a)、(c) the signal intensity of Met-6 is weak. (b)、(d) the signal of Met-6 is obvious due to the selective labeling of methionine terminal methyl group
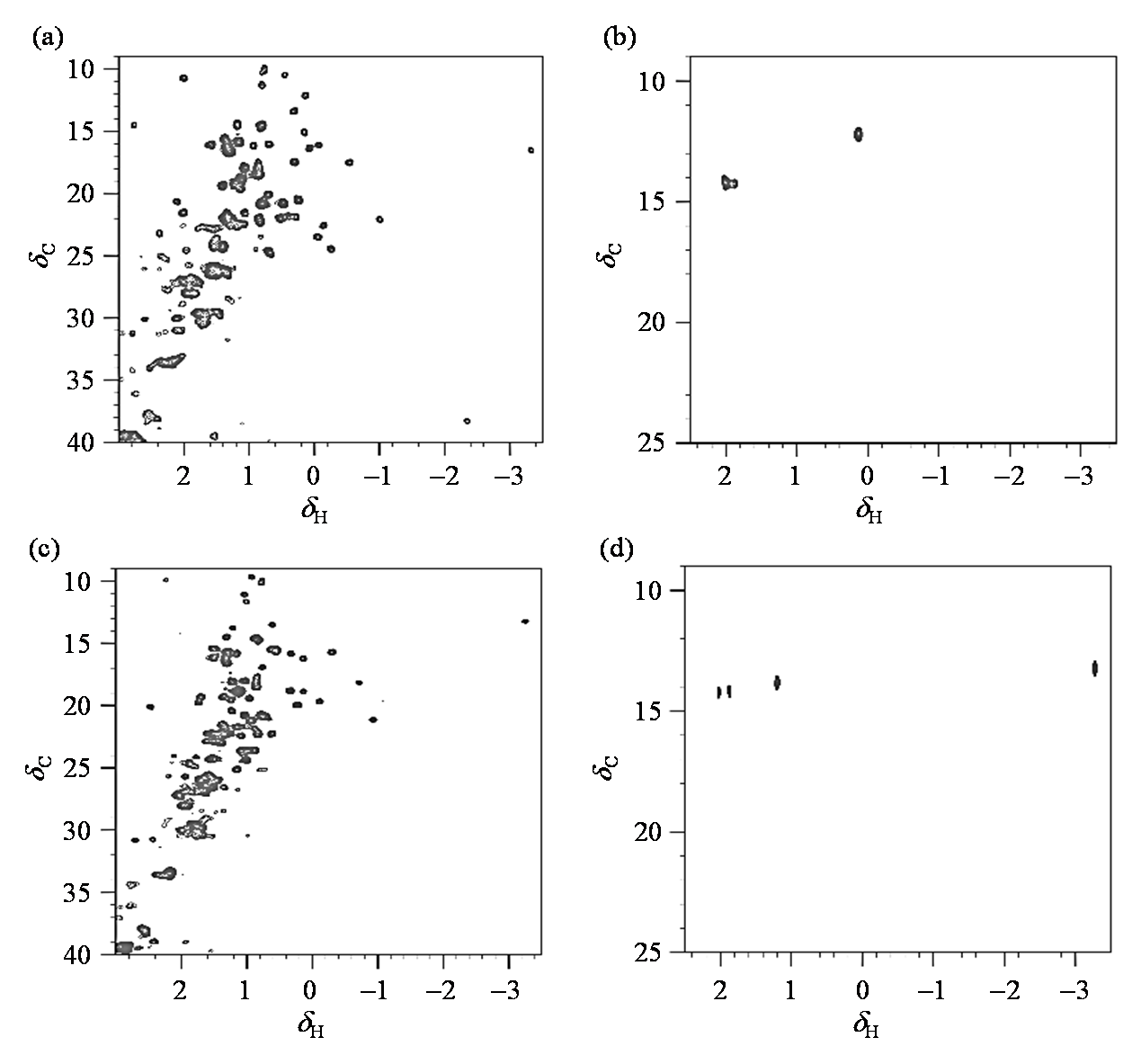
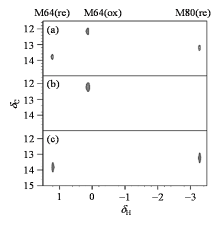
Fig. 3
1H-13C HSQC spectra of methyl-13C methionine labeled cytochrome c under different states. (a) 0.1 mmol/L methyl- 13C methionine labeled cytochrome c; (b) Sample in Fig. (a) with addition of 1 mmol/L potassium ferricyanide; (c) Sample in Fig. (a) with addition of 1 mmol/L sodium ascorbate

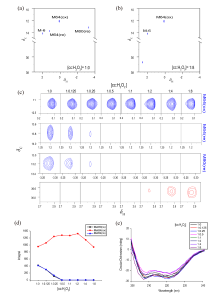
Fig. 5
1H-13C HSQC spectra and CD spectra of methyl-13C methionine labeled cytochrome c in addition with hydrogen peroxide with different concentrations. (a) 1H-13C HSQC spectrum of 25 μmol/L methyl- 13C methionine labeled cytochrome c; (b) 1H-13C HSQC spectrum of sample in Fig. (a) in addition with 200 μmol/L H2O2; (c) Methyl signals in the 1H-13C HSQC spectra of methyl-13C methionine labeled cytochrome c in addition with different concentrations of hydrogen peroxide; (d) The methionine integrals of methyl-13C methionine labeled cytochrome c change with the concentrations of added hydrogen peroxide; (e) CD spectra of methyl-13C methionine labeled cytochrome c in addition with different concentrations of hydrogen peroxide

| [1] |
SIES H. Role of metabolic H2O2 generation: redox signaling and oxidative stress[J]. J Biol Chem, 2014, 289(13): 8735-8741.
doi: 10.1074/jbc.R113.544635 |
| [2] |
KATHIRESAN M, ENGLISH A M. LC-MS/MS suggests that hole hopping in cytochrome c peroxidase protects its heme from oxidative modification by excess H2O2[J]. Chem Sci, 2017, 8(2): 1152-1162.
doi: 10.1039/C6SC03125K |
| [3] |
ZHONG F, PLETNEVA E V. Ligation and reactivity of methionine-oxidized cytochrome c[J]. Inorg Chem, 2018, 57(10): 5754-5766.
doi: 10.1021/acs.inorgchem.8b00010 pmid: 29708337 |
| [4] |
MCCALDON P, ARGOS P. Oligopeptide biases in protein sequences and their use in predicting protein coding regions in nucleotide-sequences[J]. Proteins, 1988, 4(2): 99-122.
doi: 10.1002/(ISSN)1097-0134 |
| [5] |
LUO S, LEVINE R L. Methionine in proteins defends against oxidative stress[J]. Faseb Journal, 2009, 23(2): 464-472.
doi: 10.1096/fj.08-118414 pmid: 18845767 |
| [6] |
CHAO C C, MA Y S, STADTMAN E R. Modification of protein surface hydrophobicity and methionine oxidation by oxidative systems[J]. Proc Natl Acad Sci USA, 1997, 94(7): 2969-2974.
pmid: 9096330 |
| [7] | HERSHKO A, CIECHANOVER A. The ubiquitin pathway for the degradation of intracellular proteins[J]. Prog Nucleic Acid Res Mol Biol, 1986, 33: 19-56. |
| [8] |
KEHM R, BALDENSPERGER T, RAUPBACH J, et al. Protein oxidation-formation mechanisms, detection and relevance as biomarkers in human diseases[J]. Redox Biol, 2021, 42: 101901.
doi: 10.1016/j.redox.2021.101901 |
| [9] |
LIU X S, KIM C N, YANG J, et al. Induction of apoptotic program in cell-free extracts: Requirement for dATP and cytochrome c[J]. Cell, 1996, 86(1): 147-157.
doi: 10.1016/s0092-8674(00)80085-9 pmid: 8689682 |
| [10] |
ALVAREZ-PAGGI D, HANNIBAL L, CASTRO M A, et al. Multifunctional cytochrome c: learning new tricks from an old dog[J]. Chem Rev, 2017, 117(21): 13382-13460.
doi: 10.1021/acs.chemrev.7b00257 |
| [11] |
GUERRA-CASTELLANO A, MARQUEZ I, PEREZ-MEJIAS G, et al. Post-translational modifications of cytochrome c in cell life and Disease[J]. Int J Mol Sci, 2020, 21(22): 8483
doi: 10.3390/ijms21228483 |
| [12] | SANTUCCI R, SINIBALDI F, COZZA P, et al. Cytochrome c: An extreme multifunctional protein with a key role in cell fate[J]. Int J Biol Macromol, 2019, (136): 1237-1246. |
| [13] |
KIM J, RODRIGUEZ M E, GUO M, et al. Oxidative modification of cytochrome c by singlet oxygen[J]. Free Radical Biology & Medicine, 2008, 44(9): 1700-1711.
doi: 10.1016/j.freeradbiomed.2007.12.031 |
| [14] |
WANG Z, ANDO Y, NUGRAHENI A D, et al. Self-oxidation of cytochrome c at methionine80 with molecular oxygen induced by cleavage of the Met-heme iron bond[J]. Mol Biosyst, 2014, 10(12): 3130-3137.
doi: 10.1039/c4mb00285g pmid: 25224641 |
| [15] |
IVANETICH K M, BRADSHAW J J, KAMINSKY L S. Methionine sulfoxide cytochrome-c[J]. Biochemistry, 1976, 15(5): 1144-1153.
doi: 10.1021/bi00650a029 |
| [16] |
BREN K L, RAVEN E L. Locked and loaded for apoptosis[J]. Science, 2017, 356(6344): 1236.
doi: 10.1126/science.aan5587 pmid: 28642398 |
| [17] | MUENZNER J, PLETNEVA E V. Structural transformations of cytochrome c upon interaction with cardiolipin[J]. Chem Phys Lipids, 2014, (179): 57-63. |
| [18] | TOMÁŠKOVÁ N, NOVÁK P, KOŽÁR T, et al. Early modification of cytochrome c by hydrogen peroxide triggers its fast degradation[J]. Int J Biol Macromol, 2021, (174): 413-423. |
| [19] |
YIN V, MIAN S H, KONERMANN L. Lysine carbonylation is a previously unrecognized contributor to peroxidase activation of cytochrome c by chloramine-T[J]. Chem Sci, 2019, 10(8): 2349-2359.
doi: 10.1039/C8SC03624A |
| [20] | SHI C W, SHI P, TIAN C L. NMR studies of large protein dynamics using unnatural amino acids[J]. Chinese J Magn Reson, 2021, 38(4): 523-532. |
| 史朝为, 石攀, 田长麟. 非天然氨基酸在蛋白质动态特性核磁共振研究中的应用[J]. 波谱学杂志, 2021, 38(4): 523-532. | |
| [21] | SCHÜTZ S, SPRANGERS R. Methyl TROSY spectroscopy: a versatile NMR approach to study challenging biological systems[J]. Prog Nucl Mag Res Sp, 2020, (116): 56-84. |
| [22] |
YU F, QIAO J, ROBBLEE J, et al. An integrated approach to unique NMR assignment of methionine methyl resonances in proteins[J]. Anal Chem, 2017, 89(3): 1610-1616.
doi: 10.1021/acs.analchem.6b03705 pmid: 28208280 |
| [23] |
BROOKS D J, FRESCO J R, LESK A M, et al. Evolution of amino acid frequencies in proteins over deep time: Inferred order of introduction of amino acids into the genetic code[J]. Mol Biol Evol, 2002, 19(10): 1645-1655.
pmid: 12270892 |
| [24] | LIU M, FARRANT R D, SWEATMAN B C, et al. Observation of separate J-resolved 1H NMR spectra from CH, CH2, and CH3 groups using a maximum-quantum filter[J]. J Magn Reson Ser A, 1995, (1064-1858): 251-256. |
| [25] | FANG Z P, SUN P, WANG Q W, et al. Conformational change of wild type cytochrome c characterized by NMR spectroscopy at natural isotropic abundance[J]. Chinese J Magn Reson, 2019, 36(4): 481-489. |
| 方仲佩, 孙鹏, 王倩文, 等. 天然同位素丰度野生型酵母细胞色素c构象变化的核磁共振检测[J]. 2019, 36(4): 481-489. | |
| [26] |
TURNER H S A D L. 13C and proton NMR studies of horse cytochrome c assignment and temperature dependence of methyl resonances[J]. Federation of European Biochemical Societies, 1986, 194(3116): 73-77.
doi: 10.1016/0014-5793(86)80054-0 |
| [27] |
HIREL PH, SCHMITTER J-M, DESSEN P, et al. Extent of N-terminal methionine excision from Escherichia coli proteins is governed by the side-chain length of the penultimate amino acid[J]. Proc Natl Acad Sci USA, 1989, 86(21): 8247-8251.
pmid: 2682640 |
| [28] |
SUN P, WANG Q, YUAN B, et al. Monitoring alkaline transitions of yeast iso-1 cytochrome c at natural isotopic abundance using trimethyllysine as a native NMR probe[J]. Chem Commun (Camb), 2018, 54(89): 12630-12633.
doi: 10.1039/c8cc07605g pmid: 30351312 |
| [29] |
VOLKOV A, WORRALL J, HOLTZMANN E, et al. Solution structure and dynamics of the complex between cytochrome c and cytochrome c peroxidase determined by paramagnetic NMR[J]. Proc Natl Acad Sci USA, 2006, 103(50): 18945-18950.
pmid: 17146057 |
| [30] |
PARAKRA R D, KLEFFMANN T, JAMESON G N L, et al. The proportion of Met80-sulfoxide dictates peroxidase activity of human cytochrome c[J]. Dalton Trans, 2018, 47(27): 9128-9135.
doi: 10.1039/c8dt02185f pmid: 29944150 |
| [31] | BERGHUIS A M B, G. D. Oxidation state-dependent conformational changes in cytochrome c[J]. J Mol Biol, 1992, (223): 959-976. |
| [32] | BUSHNELL G W, LOUIE G V, BRAYER G D. High-resolution 3-dimensional structure of horse heart cytochrome c[J]. J Mol Biol, 1990, (214): 585-595. |
| [1] | ZHAN Jianhua,HU Qin,ZHU Qinjun,JIANG Bin,ZHANG Xu,LIU Maili. Track the Conformational Change of Unlabeled Yeast Cytochrome c in Cell Homogenate Using NMR [J]. Chinese Journal of Magnetic Resonance, 2023, 40(1): 22-29. |
| [2] | Shu-huai ZHANG,Hui MA,Zhao-hui GUO,Min-jun MA,Yan QIAO,Ying-xiong WANG. Interactions Between n-Butanol/Propionic Acid and Humic Acid Studied by NMR Spectroscopy [J]. Chinese Journal of Magnetic Resonance, 2021, 38(3): 301-312. |
| [3] | LI Yu-jiang, ZHAO Wei, GUO Xiao-he, TAO Le, ZHANG Xiang, ZHANG Hai-yan, ZHAO Tian-zeng. NMR Data Analysis of Manidipine Hydrochloride [J]. Chinese Journal of Magnetic Resonance, 2021, 38(1): 110-117. |
| [4] | WANG Si-hong, ZHANG Jing-dong, YIN Xiu-mei, LI Dong-hao, KAN Yu-he, HU Wei. NMR Assignments of 6-(4-chlorophenoxy)-tetrazolo[5,1-a]phthalazine [J]. Chinese Journal of Magnetic Resonance, 2020, 37(3): 390-398. |
| [5] | FANG Zhong-pei, SUN Peng, WANG Qian-wen, ZHANG Liang, LIU Mai-li, ZHANG Xu. Conformational Change of Wild Type Cytochrome c Characterized by NMR Spectroscopy at Natural Isotropic Abundance [J]. Chinese Journal of Magnetic Resonance, 2019, 36(4): 481-489. |
| [6] | NING Cai-fang, MA Min-jun, GUO Zhao-hui, ZHANG Shu-huai, QIAO Yan, WANG Ying-xiong. Interactions Between 5-Fluorouracil and PAMAM Dendrimers Studies by NMR Spectroscopy [J]. Chinese Journal of Magnetic Resonance, 2019, 36(4): 555-562. |
| [7] | LIU Wen-qing, SONG Yan-hong, WANG Xue-lu, YAO Ye-feng. In Operando Nuclear Magnetic Resonance Spectroscopy Study on Photocatalytic Methanol Reforming [J]. Chinese Journal of Magnetic Resonance, 2019, 36(3): 298-308. |
| [8] | LI Hong-wei, YUAN Zhi-liang, XIA Bin. Determination of Apparent Protein Molecular Weight in Solution by Diffusion Ordered NMR Spectroscopy [J]. Chinese Journal of Magnetic Resonance, 2018, 35(3): 280-286. |
| [9] | HUANG Jun-lin, YU Yi-hua. Effects of Digital Resolution on Diffusional Dimension in DOSY Experiments [J]. Chinese Journal of Magnetic Resonance, 2018, 35(3): 287-293. |
| [10] | GUO Hai-qing, XIN Jia-xiang, LIU Hui-xia, WEI Da-xiu, YAO Ye-feng. Preparation of Long-Lived Nuclear Singlet States in Three-Spin Systems [J]. Chinese Journal of Magnetic Resonance, 2018, 35(3): 345-352. |
| [11] | ZHOU Zhong-gao, YUAN Yang-yang, LIU Hong-bo, QI Qi, XIE Yong-rong. An NMR Study on Prucalopride [J]. Chinese Journal of Magnetic Resonance, 2018, 35(1): 119-127. |
| [12] | SUN Wei, SHE Meng-yao, ZONG Chun-lei, GAO Xiang, GUO Juan. An NMR Study on Diacetonefructose [J]. Chinese Journal of Magnetic Resonance, 2017, 34(3): 329-337. |
| [13] | ZHAI Zi-ning, WU Qiong, LI Cong-gang. Lysine Acetylation Inhibits α-Synuclein Fibrillation [J]. Chinese Journal of Magnetic Resonance, 2016, 33(2): 179-187. |
| [14] | LIU De-yun, WANG Ya-ru, WEI Hai-feng, XIA Gao-feng, YANG Xiao-yun. Spectral Analysis of Fungicide Cyflufenamid [J]. Chinese Journal of Magnetic Resonance, 2016, 33(1): 142-152. |
| [15] |
PANG Zi-bo*, MENG Da-lei, DOU Ying, MA Si-rui, WU Cong,CHENG Hong-juan, XU Yong-kuan.
Characterization of Molecular Structure of DAST Using Nuclear Magnetic Resonance Spectroscopy [J]. Chinese Journal of Magnetic Resonance, 2015, 32(4): 637-647. |
| Viewed | ||||||||||||||||||||||||||||||||||||||||||||||||||
|
Full text 492
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||
|
Abstract 235
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||