引言
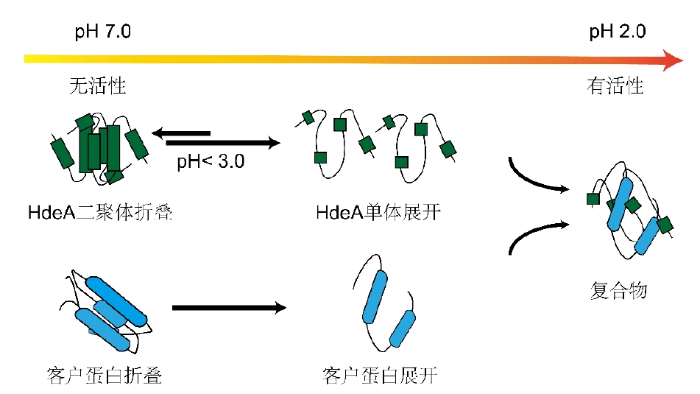
细菌周质是指革兰氏阴性菌中处于内膜与外膜之间的狭窄空间,其内部包含了许多周质蛋白、核酸、脂类以及代谢物[1].HdeA是一种定位在细菌周质的分子伴侣,对维持细菌周质中蛋白质稳态发挥着重要功能[2],当肠道细菌通过宿主的胃时,其周质环境处于酸性,导致周质蛋白变性引发聚集.此时周质分子伴侣HdeA迅速从同源二聚体解离成单体,激活其分子伴侣活性,与天然客户蛋白包括外膜蛋白(Outer membrane proteins,OMPs)以及其它伴侣蛋白SurA和Degp等[3,4]相互作用.单体结构部分展开并暴露出疏水表面,通过与底物蛋白结合阻止其酸诱导的聚集.当细菌移动到小肠时,此时处于中性环境,HdeA又释放其客户蛋白,并重新折叠成同源二聚体形式,此时不具备分子伴侣活性.其构象变化的示意图如图1所示.了解HdeA如何进行酸激活伴侣活性,有助于更好地理解各类消化道细菌的生理功能.
图1
细菌外膜囊泡(Outer membrane vesicles,OMVs)是革兰氏阴性菌自发分泌的天然球型囊泡,其内部充满了周质可溶物,具有与细菌周质极为相似的天然环境,是作为研究周质蛋白和外膜蛋白的理想原位环境[9,10].相比较于活细胞体系,它的无生命特性能够支持长时间采样的NMR研究,同时其较强的耐酸碱性提供了比较广泛的pH研究范围.在本研究中,通过在HdeA的N端融合分泌信号肽,将大量的HdeA过表达在细菌周质中,当HdeA被随机包裹于OMVs中后,分泌至培养基中,经过高速离心法获取富含HdeA的OMVs样品.利用液体NMR技术进一步表征了稀溶液体系和OMVs体系中pH对HdeA的结构和构象变化的影响.
1 实验部分
1.1 仪器与试剂
1.1.1 菌株
实验采用的菌株为BL21(DE3)大肠杆菌(E. coli).
1.1.2 药品与试剂
磷酸氢二钠(Na2HPO4)、磷酸二氢钠(NaH2PO4)、磷酸二氢钾(KH2PO4)、氯化钠(NaCl)、氢氧化钠(NaOH)、氯化钙(CaCl2)、硫酸钾(K2SO4)、甘油(Glycerol)、柠檬酸钠(Sodium citrate)等分析级化学品购自国药试剂.蛋白胨(Tryptone)、酵母粉(Yeast extract)购自Oxiod.苯甲基磺酰氟(PMSF)、异丙基硫代半乳糖苷(IPTG)购自碧云天生物.FastHifi Super DNA聚合酶预混液购自Affinibody Life Science AG.
1.1.3 仪器
实验仪器包括:600 MHz 核磁共振波谱仪(Bruker,AVANCE III 600MHz,德国),蛋白纯化仪(GE HealthCare,AKTA prime plus,美国).
1.2 样品制备
1.2.1 HdeA-OMVs样品的制备
HdeA-OMVs样品的制备方法参考已发表的文献[9],用M9培养基对HdeA进行同位素标记,由于在HdeA的N端融合了分泌信号肽,在诱导HdeA表达的过程中,HdeA被随机的包裹在OMVs中并分泌到培养基中.具体步骤如下:(1)将诱导完成的1 L菌液经过离心(6 000g,30 min,4 ℃)得到培养基上清.(2)将大约1 L上清经过100 kDa的膜包浓缩至20 mL.(3)将所得浓缩液经过0.45 μm的滤膜过滤之后,再进行离心(40 000g,3 h,4 ℃),弃去上清.(4)加入1.5 mL磷酸缓冲液(20 mmol/L PB,150 mmol/L NaCl,pH 6.5)进行重悬,经过离心(1 000g,1 min,4 ℃),小心的吸取乳浊液,弃去不溶性沉淀,重复数次.(5)将乳浊液进行离心(20 000g,1 h,4 ℃),沉淀即为OMVs,最后再用PBS缓冲液进行重悬,得到HdeA-OMVs样品.
1.2.2 HdeA的分离和纯化
HdeA的分离和纯化步骤:(1)将诱导完成的菌液经过离心(6 000g,30 min,4 ℃)得到培养基上清. (2)将1 L上清经过100 kDa的膜包浓缩至20 mL.(3)将浓缩后的样品经过3.5 kDa透析膜的在Buffer A(10 mmol/L PB,pH 6.5)中透析两次,每次透析4 h.(4)然后将样品经过强阴离子交换柱(HiTrap Q FF)进行初步纯化,用Buffer B(20 mmol/L PB,500 mmol/L NaCl,pH 6.5)进行梯度洗脱.(5)最后经过尺寸排阻色谱(SEC)分离纯化,将蛋白以合适浓度保存在Buffer C中(20 mmol/L PB,150 mmol/L NaCl,pH 6.5).
1.2.3 NMR样品的制备
本研究中涉及的相同pH条件下的HdeA稀溶液样品(100 μmol/L HdeA)与HdeA-OMVs样品在同一种缓冲液(20 mmol/L柠檬酸钠,150 mmol/L NaCl,pH 6.5或pH 2.5)中透析过夜,以保证HdeA稀溶液样品和HdeA-OMVs样品保持相同pH值.OMVs是由细菌的外膜所形成的囊泡,其表面存在许多跨膜孔蛋白,允许600 Da以下的水溶性小分子和离子以扩散的形式自由进出,通过透析的方式可以达到调节OMVs内腔pH的目的[11⇓-13].pH 3.5、pH 4.5及pH 5.5的稀溶液样品分别通过在对应pH的缓冲液(20 mmol/L柠檬酸钠,150 mmol/L NaCl)中透析过夜获得.相关实验操作参考已发表的文献[9,14].
1.3 磁共振波谱采集与数据处理
[15N,1H]-HSQC实验在配备了三共振低温探头的Bruker 600 MHz核磁共振谱仪上采集,实验温度设置为298 K,所有实验均采用标准脉冲程序.1H和15N的工作频率分别为600.17 MHz和60.82 MHz,在1H 和15N维的谱宽分别为9 615.38 Hz和2 189.30 Hz,谱中心分别为2 809.70 Hz和7 175.66 Hz,采样数据点阵t2×t1 = 2 048×96,弛豫等待时间d1 = 1.5 s,累加次数为8,对于HdeA-OMVs样品,累加次数为64.实验数据通过Bruker Topspin 4.0.1软件以及CcpNmr软件[15]进行处理.
2 结果与讨论
2.1 HdeA蛋白在OMVs中的富集
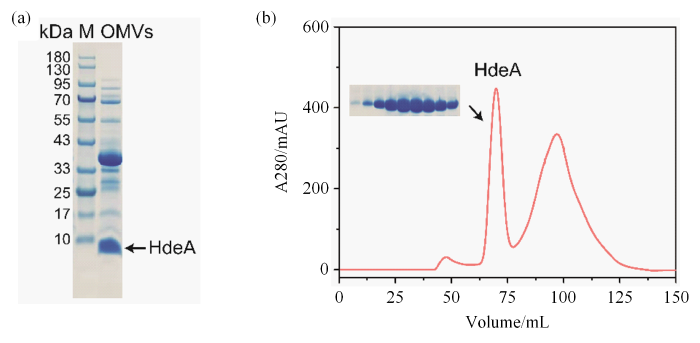
在先前的研究中,对OMVs蛋白定量分析表明,E. coli可以将大约0.2%~0.5%的外膜和周质蛋白包裹到OMVs中[16⇓-18].这些蛋白质包括毒素、黏附素、毒力因子、蛋白酶、糖蛋白等,在营养获取、生物膜的形成、压力响应、对宿主的入侵和免疫调节等过程中发挥重要的作用[19,20].尽管现在对OMVs包裹周质蛋白的选择机制尚不清楚,但是采用过表达的方式富集目的蛋白到OMVs中是一种理想的策略.我们通过浓缩和超速离心结合的方式,大量制备HdeA-OMVs样品.如图2(a)所示,HdeA-OMVs样品在SDS-PAGE电泳胶图上清晰可见HdeA条带,表明HdeA经过诱导表达后,被大量富集在OMVs腔内.另外,电泳胶图上还存在除HdeA之外的高浓度蛋白条带,通过胶条质谱鉴定以及结合参考文献报道[10],这些条带被鉴定为膜蛋白.此外培养液上清中也分泌了大量的HdeA,对此部分蛋白进行纯化,获得了稀溶液中测量的HdeA蛋白样品,用于后续实验.图2(b)表示HdeA在经过SEC纯化后得到的色谱图以及SDS-PAGE电泳胶图.
图2
图2
(a) HdeA-OMVs样品在SDS-PAGE电泳胶图,箭头标注条带(~10 kDa)为HdeA,其余条带为OMVs中的周质蛋白或外膜蛋白;(b)培养基上清中的HdeA蛋白经过SEC分离后的液相色谱图,在图中约68 mL的位置为HdeA的二体峰
Fig. 2
(a) SDS-PAGE gel plot of HdeA-OMVs samples, the arrow marked band (~10 kDa) is HdeA, and the remaining bands are pericytoplasmic proteins or outer membrane proteins in OMVs; (b) Liquid chromatogram of the HdeA from medium supernatant separated after size exclusion chromatography, in which the peak of the dimer HdeA is located at about 68 mL
2.2 HdeA的[15N,1H]-HSQC谱图
2.2.1 HdeA在稀溶液条件下的[15N,1H]-HSQC谱图
图3
图3
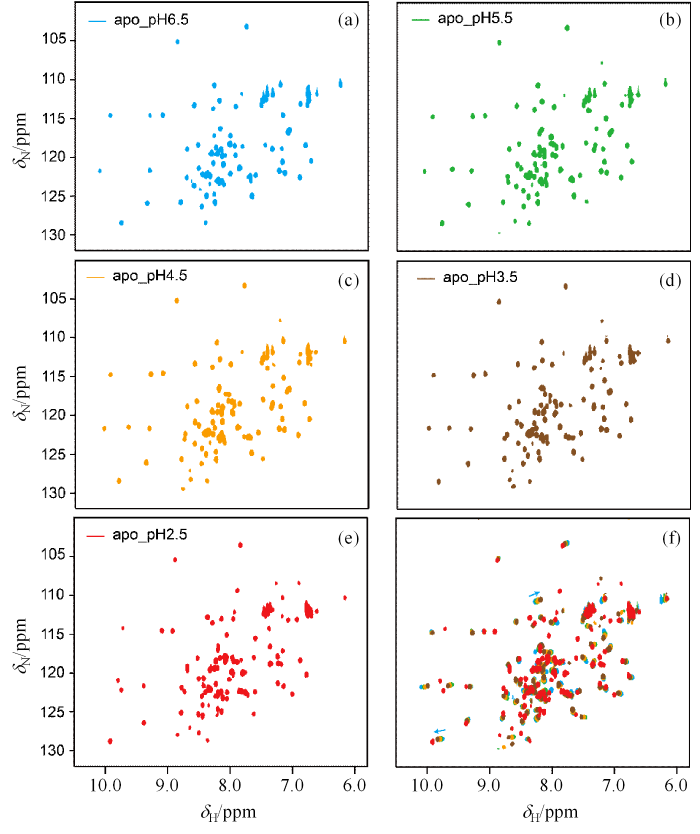
HdeA在不同pH稀溶液中的[15N,1H]-HSQC谱图. (a) pH 6.5(蓝色);(b) pH 5.5(绿色);(c) pH 4.5(黄色);(d) pH 3.5(棕色);(e) pH 2.5(红色);(f) HdeA在不同pH溶液中[15N,1H]-HSQC谱的对比图
Fig. 3
The [15N,1H]-HSQC spectra of HdeA in buffers with different pH. (a) pH 6.5 (blue); (b) pH 5.5 (green); (c) pH 4.5 (yellow); (d) pH 3.5 (brown); (e) pH 2.5 (red); (f) Comparison of [15N,1H]-HSQC spectra of HdeA in buffers with different pH
从谱图中分析可得,在pH 2.5条件下HdeA主链H-N谱峰数目相较于pH 6.5条件下的谱图主链H-N谱峰数,从68个增加到89个,表明HdeA的构象发生了变化.且HdeA共89个氨基酸,除4个脯氨酸和第一个丙氨酸外,理论上最多在NMR谱图中观察到84个主链H-N信号. 多的5个H-N谱峰信号,进一步表明HdeA在pH 2.5的条件下,处于两态或者多态的慢交换中.根据之前的参考文献[3],HdeA在pH 2.5环境中可能处于二体与单体的交换,或者存在折叠的HdeA与去折叠的HdeA之间的交换.
2.2.2 HdeA在OMVs腔内的[15N,1H]-HSQC谱图
图4
图4
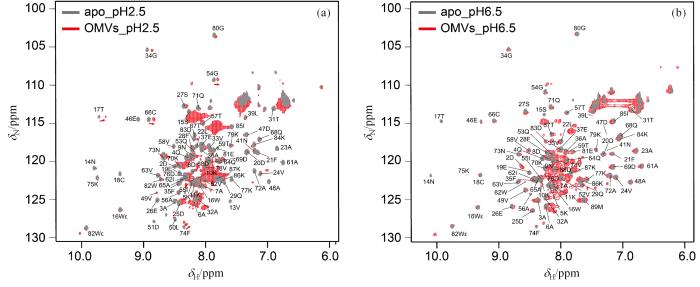
HdeA在稀溶液(灰色)和OMVs(红色)中的[15N, 1H]-HSQC谱图(a)pH 2.5;(b) pH 6.5
Fig. 4
The [15N, 1H]-HSQC spectra of HdeA in the buffer (grey) and in OMVs (red) at (a) pH 2.5 and (b) pH 6.5
对于分散峰,在pH 6.5条件下,HdeA在稀溶液以及OMVs中的谱图,其主链残基基本一致,表明其总体结构基本一致,同时也暗示HdeA并未与OMVs中的其它生物大分子发生相互作用.在pH 2.5条件下,HdeA在稀溶液和OMVs中的[15N,1H]-HSQC谱图显示在HdeA-OMVs样品中有部分HdeA残基发生了明显化学位移扰动,这说明在pH 2.5条件下稀溶液中和OMVs拥挤环境下的HdeA存在一定的差别.
(1)式中,ΔH和ΔN分别表示HdeA残基在1H以及15N维的化学位移扰动差值.
图5
图5
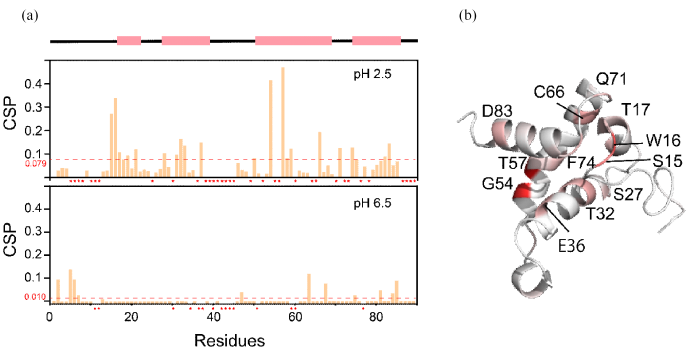
HdeA在OMVs中残基的化学位移扰动.(a) pH 2.5和pH 6.5条件下,HdeA在OMVs环境中与在稀溶液中各残基的CSP,其中*号表示该残基由于峰重叠导致未能得到可信的CSP数据;(b) HdeA的单体结构,红色标注部分为CSP值变化较大的残基
Fig. 5
The CSP of residues of HdeA in OMVs. (a) Chemical shift perturbation difference of each residue of HdeA in the OMVs environment and dilute solution at pH 2.5 and pH 6.5, where * indicates that no credible CSP data could be obtained for this residue due to peak overlap; (b) Monomer structure of HdeA, in which residues with large variation in CSP values are marked in red
图5结果显示在pH 2.5条件下,HdeA在OMVs原位环境中与在稀溶液中相比,各分散峰的残基平均CSP为0.079,大于其在pH 6.5条件下OMVs环境中与稀溶液环境中的平均CSP 0.010.在pH 2.5条件下,相比于稀溶液条件,OMVs中HdeA上S15、W16、T17、S27、T32、E36、G54、T57、C66、Q71、F74以及D83等残基表现出较大的化学位移扰动.这些残基的化学位移扰动较大可能由以下几个原因引起:(1)OMVs中HdeA在pH 2.5条件下结构发生变化,部分残基暴露后可与天然周质环境中的客户蛋白发生相互作用;(2)OMVs中HdeA局部浓度高于稀溶液中的HdeA浓度,导致两种环境中的HdeA在pH 2.5条件下最先发生部分去折叠的区域及去折叠路径不同,从而引起显著的化学位移扰动差异.如上结果揭示了HdeA在原位环境中激活其分子伴侣活性的可能分子机制.
3 结论
在本研究中,通过[15N,1H]-HSQC实验对OMVs中分子伴侣HdeA进行了初步表征.首先,在不同pH条件下分别采集了稀溶液及OMVs中的HdeA [15N,1H]-HSQC谱图.结果显示在OMVs中pH 6.5及pH 2.5条件下均获得了高分辨率的HdeA [15N,1H]-HSQC谱图.我们观察到HdeA在OMVs中的[15N,1H]-HSQC谱图上同时存在聚集峰和分散峰,其中聚集峰分布在δH 8.0附近,谱峰重叠严重导致难以归属,推测其主要来源于OMVs中HdeA及其它蛋白的柔性或无结构区域.在已有文献基础上[4],我们对分散峰进行了归属及进一步的研究,揭示了天然OMVs环境中HdeA在酸刺激下产生与稀溶液环境中不同的构象变化.这些为后续更深入的研究奠定了基础.同时与同质环境相似的天然OMVs为后续原位研究分子伴侣-客户蛋白相互作用及功能机制提供了新的体系.
致谢
感谢国家重点基础研究发展计划(2017YFA0505400,2018YFE0202300,2018YFA0704002)的支持,国家自然科学基金资助项目(22174151,21904138,21991080,22078280)的支持.
利益冲突
无
参考文献
Protein folding in the periplasm of Escherichia coli
[J].DOI:10.1111/mmi.1994.12.issue-5 URL [本文引用: 1]
Chaperone-dependent mechanisms for acid resistance in enteric bacteria
[J].
DOI:10.1016/j.tim.2012.03.001
PMID:22459131
[本文引用: 1]

The extremely acidic environment of the mammalian stomach not only serves to facilitate food digestion but also acts as a natural barrier against infections of food-borne pathogens. Many pathogenic bacteria, such as enterohemorrhagic Escherichia coli, can breach this host defense and cause severe diseases. These pathogens have evolved multiple intricate strategies to overcome the bactericidal activity of acids. In particular, recent studies have uncovered the central roles of two periplasmic chaperones, HdeA and HdeB, in protecting enteric bacteria from extremely acidic conditions. Here, we review recent advances in the understanding of the acid resistance mechanisms of Gram-negative bacteria and focus on the mechanisms of HdeA and HdeB in preventing acid-induced protein aggregation and facilitating protein refolding following pH neutralization.Copyright © 2012 Elsevier Ltd. All rights reserved.
Structural basis and mechanism of the unfolding-induced activation of HdeA, a bacterial acid response chaperone
[J].DOI:10.1074/jbc.RA118.006398 URL [本文引用: 7]
NMR-monitored titration of acid-stress bacterial chaperone HdeA reveals that Asp and Glu charge neutralization produces a loosened dimer structure in preparation for protein unfolding and chaperone activation
[J].
DOI:10.1002/pro.2402
PMID:24375557
[本文引用: 3]

HdeA is a periplasmic chaperone found in several gram-negative pathogenic bacteria that are linked to millions of cases of dysentery per year worldwide. After the protein becomes activated at low pH, it can bind to other periplasmic proteins, protecting them from aggregation when the bacteria travel through the stomach on their way to colonize the intestines. It has been argued that one of the major driving forces for HdeA activation is the protonation of aspartate and glutamate side chains. The goal for this study, therefore, was to investigate, at the atomic level, the structural impact of this charge neutralization on HdeA during the transition from near-neutral conditions to pH 3.0, in preparation for unfolding and activation of its chaperone capabilities. NMR spectroscopy was used to measure pKa values of Asp and Glu residues and monitor chemical shift changes. Measurements of R2/R1 ratios from relaxation experiments confirm that the protein maintains its dimer structure between pH 6.0 and 3.0. However, calculated correlation times and changes in amide protection from hydrogen/deuterium exchange experiments provide evidence for a loosening of the tertiary and quaternary structures of HdeA; in particular, the data indicate that the dimer structure becomes progressively weakened as the pH decreases. Taken together, these results provide insight into the process by which HdeA is primed to unfold and carry out its chaperone duties below pH 3.0, and it also demonstrates that neutralization of aspartate and glutamate residues is not likely to be the sole trigger for HdeA dissociation and unfolding.© 2013 The Protein Society.
Track the conformational change of unlabeled yeast cytochrome c in cell homogenate using NMR
[J].
基于磁共振的胞浆中无标记酵母细胞色素c构象变化追踪
[J].
Characterizations of the interactions between Escherichia coli periplasmic chaperone HdeA and its native substrates during acid stress
[J].DOI:10.1021/acs.biochem.7b00724 URL [本文引用: 3]
The mechanism of HdeA unfolding and chaperone activation
[J].
DOI:S0022-2836(17)30540-5
PMID:29138002
[本文引用: 1]

HdeA is a periplasmic chaperone that is rapidly activated upon shifting the pH to acidic conditions. This activation is thought to involve monomerization of HdeA. There is evidence that monomerization and partial unfolding allow the chaperone to bind to proteins denatured by low pH, thereby protecting them from aggregation. We analyzed the acid-induced unfolding of HdeA using NMR spectroscopy and fluorescence measurements, and obtained experimental evidence suggesting a complex mechanism in HdeA's acid-induced unfolding pathway, as previously postulated from molecular dynamics simulations. Counterintuitively, dissociation constant measurements show a stabilization of the HdeA dimer upon exposure to mildly acidic conditions. We provide experimental evidence that protonation of Glu37, a glutamate residue embedded in a hydrophobic pocket of HdeA, is important in controlling HdeA stabilization and thus the acid activation of this chaperone. Our data also reveal a sharp transition from folded dimer to unfolded monomer between pH3 and pH 2, and suggest the existence of a low-populated, partially folded intermediate that could assist in chaperone activation or function. Overall, this study provides a detailed experimental investigation into the mechanism by which HdeA unfolds and activates.Copyright © 2017 Elsevier Ltd. All rights reserved.
Roles of structural plasticity in chaperone HdeA activity are revealed by 19F NMR
[J].DOI:10.1039/C5SC04297F URL [本文引用: 1]
Protein conformational exchanges modulated by the environment of outer membrane vesicles
[J].
DOI:10.1021/acs.jpclett.3c00152
PMID:36897994
[本文引用: 3]

Protein function, in many cases, is strongly coupled to the dynamics and conformational equilibria of the protein. The environment surrounding proteins is critical for their dynamics and can dramatically affect the conformational equilibria and subsequently the activities of proteins. However, it is unclear how protein conformational equilibria are modulated by their crowded native environments. Here we reveal that outer membrane vesicle (OMV) environments modulate the conformational exchanges of Im7 protein at its local frustrated sites and shift the conformation toward its ground state. Further experiments show both macromolecular crowding and quinary interactions with the periplasmic components stabilize the ground state of Im7. Our study highlights the key role that the OMV environment plays in the protein conformational equilibria and subsequently the conformation-related protein functions. Furthermore, the long-lasting nuclear magnetic resonance measurement time of proteins within OMVs indicates that they could serve as a promising system for investigating protein structures and dynamics via nuclear magnetic spectroscopy.
Protein-enriched outer membrane vesicles as a native platform for outer membrane protein studies
[J].
DOI:10.1038/s42003-018-0027-5
[本文引用: 2]

Most studies characterizing the folding, structure, and function of membrane proteins rely on solubilized or reconstituted samples. Whereas solubilized membrane proteins lack the functionally important lipid membrane, reconstitution embeds them into artificial lipid bilayers, which lack characteristic features of cellular membranes including lipid diversity, composition and asymmetry. Here, we utilize outer membrane vesicles (OMVs) released from Escherichia coli to study outer membrane proteins (Omps) in the native membrane environment. Enriched in the native membrane of the OMV we characterize the assembly, folding, and structure of OmpG, FhuA, Tsx, and BamA. Comparing Omps in OMVs to those reconstituted into artificial lipid membranes, we observe different unfolding pathways for some Omps. This observation highlights the importance of the native membrane environment to maintain the native structure and function relationship of Omps. Our fast and easy approach paves the way for functional and structural studies of Omps in the native membrane.
Molecular basis of bacterial outer membrane permeability
[J].DOI:10.1128/mr.49.1.1-32.1985 PMID:2580220 [本文引用: 1]
General and specific porins from bacterial outer membranes
[J].Over the past years, the three-dimensional structures of several bacterial porins have been determined to high resolution. Apart from revealing an unusual type of architecture, the hollow beta-barrel, they have made it possible to investigate in detail various structure-function relationships. Characteristics of ion flow through (native and modified) porins inserted into artificial bilayers have been related to the electrostatic properties of the pores. The structural basis of voltage induced pore closing, however, is still not resolved. The remarkable ability of maltoporin to allow translocation of long maltodextrin molecules through the small channel has been traced back to the presence of an elongated hydrophobic patch at the channel lining.
Structure and function of bacterial outer membrane proteins: barrels in a nutshell
[J].
DOI:10.1046/j.1365-2958.2000.01983.x
PMID:10931321
[本文引用: 1]

The outer membrane protects Gram-negative bacteria against a harsh environment. At the same time, the embedded proteins fulfil a number of tasks that are crucial to the bacterial cell, such as solute and protein translocation, as well as signal transduction. Unlike membrane proteins from all other sources, integral outer membrane proteins do not consist of transmembrane alpha-helices, but instead fold into antiparallel beta-barrels. Over recent years, the atomic structures of several outer membrane proteins, belonging to six families, have been determined. They include the OmpA membrane domain, the OmpX protein, phospholipase A, general porins (OmpF, PhoE), substrate-specific porins (LamB, ScrY) and the TonB-dependent iron siderophore transporters FhuA and FepA. These crystallographic studies have yielded invaluable insight into and decisively advanced the understanding of the functions of these intriguing proteins. Our review is aimed at discussing their common principles and peculiarities as well as open questions associated with them.
High-resolution in situ NMR spectroscopy of bacterial envelope proteins in outer membrane vesicles
[J].DOI:10.1021/acs.biochem.9b01123 URL [本文引用: 2]
The CCPN data model for NMR spectroscopy: development of a software pipeline
[J].DOI:10.1002/prot.v59:4 URL [本文引用: 1]
Release of outer membrane fragments from normally growing Escherichia coli
[J].
Preferential release of new outer membrane fragments by exponentially growing Escherichia coli
[J].
Incorporation of heterologous outer membrane and periplasmic proteins into Escherichia coli outer membrane vesicles
[J].DOI:10.1074/jbc.M307628200 URL [本文引用: 1]
Selective sorting of cargo proteins into bacterial membrane vesicles
[J].
DOI:10.1074/jbc.M110.185744
PMID:21056982
[本文引用: 1]

In contrast to the well established multiple cellular roles of membrane vesicles in eukaryotic cell biology, outer membrane vesicles (OMV) produced via blebbing of prokaryotic membranes have frequently been regarded as cell debris or microscopy artifacts. Increasingly, however, bacterial membrane vesicles are thought to play a role in microbial virulence, although it remains to be determined whether OMV result from a directed process or from passive disintegration of the outer membrane. Here we establish that the human oral pathogen Porphyromonas gingivalis has a mechanism to selectively sort proteins into OMV, resulting in the preferential packaging of virulence factors into OMV and the exclusion of abundant outer membrane proteins from the protein cargo. Furthermore, we show a critical role for lipopolysaccharide in directing this sorting mechanism. The existence of a process to package specific virulence factors into OMV may significantly alter our current understanding of host-pathogen interactions.
Protein selection and export via outer membrane vesicles
[J].
Characterization of residue specific protein folding and unfolding dynamics in cells
[J].
DOI:10.1021/jacs.9b04435
PMID:31305080
[本文引用: 1]

In this work, we measured the millisecond residue specific protein folding and unfolding dynamics in cells for two protein GB3 mutants using NMR. The results show that the protein folding and unfolding dynamics in cells is different from that in buffer. Through a two-site exchange model, it is shown that both the population and the exchange rate are changed by the cellular environment. Further investigation suggests that the change is likely due to the quinary interaction with crowded molecules in the cell. Our work underlines the importance of cellular environment to protein folding kinetics and thermodynamics although this environmental effect may not be large enough to change the protein structure.
Quantitative comparison of protein dynamics in live cells and in vitro by in-cell 19F NMR
[J].DOI:10.1039/c3cc39205h URL [本文引用: 1]
Using chemical shift perturbation to characterize ligand binding
[J].DOI:10.1016/j.pnmrs.2013.02.001 URL [本文引用: 1]