引言
《中国心血管病报告2018》概要表明,心血管病(cardiovascular disease,CVDs)死亡率居于我国居民死亡率首位,其占比达到40%以上,且CVDs患病率和死亡率仍在持续上升[1].因此,迫切需要能提供心脏形态和功能定量信息的技术,以辅助诊断心脏疾病.心脏磁共振电影成像(cine cardiac magnetic resonance imaging,Cine-CMRI)技术无电离辐射,时空分辨率高,可重复性好[2].它采用稳态自由进动(steady-state free precession,SSFP)序列得到心肌和血液对比度更高的亮血图像[3],可完整清楚地显示心室和心肌,被广泛应用于临床心室心肌分割与功能评估中.
左心室(left ventricle,LV)是人体血液循环的泵体.左心肌很厚,具有足够的动力将富含氧和营养物质的动脉血泵向全身.基于Cine-CMRI的左心肌分割,可计算心室体积、心肌质量等功能指标,对诊断心脏疾病(如心肌缺血、心肌梗死等)具有重要意义.准确的左心肌分割是评估心脏功能指标的基础,通常由专家手动勾画实现,繁琐耗时,受主观因素影响大.因此,大批研究人员着力于研究左心肌分割,不断提出各分割方法,实现左心肌快速准确地分割.然而,左心肌与周围组织灰度呈现一致性,且乳头肌、小梁肌常与LV内膜相连,给左心肌分割带来一定的难度[4].
迄今,研究者已提出多种基于Cine-CMRI的左心肌分割方法,即分割LV内外膜轮廓.传统分割算法主要包括区域生长法、动态规划、水平集法、主动轮廓模型等.Santiago等[5]提出了一种结合LV中心和半径先验知识的动态规划法,将MR图像转换到极坐标下提取LV内外膜轮廓.Yang等[6]通过圆形约束的距离正则化水平集演化模型分割LV内膜,并利用区域可伸缩拟合模型提取LV外膜.Lee等[7]将迭代阈值法分割得到的LV内膜作为主动轮廓模型的初始化,结合图像强度约束提取LV外膜.这些方法对初始位置和图像质量敏感,且需要广泛的用户介入,不能兼顾计算复杂度和分割结果的准确性、鲁棒性.近年来,人工神经网络不断发展,深度学习兴起并被广泛运用至模式识别、图像分类、语义分割等领域.继2015年Kaggle第二届年度数据科学挑战赛后,大量深度学习的方法被应用于CMRI心脏分割[8].Avendi等[9]结合深度学习与传统方法来分割LV,首先利用卷积神经网络(convolutional neural network,CNN)定位LV,再通过堆叠自编码器推测LV形状,将其当作形变模型的初始化展开分割,克服了因乳头肌存在、LV顶部分辨率低导致边界分割不准确的问题.此外,Ngo等[10]、Yang等[11]也提出将深度学习与传统方法结合分割左心肌,提高分割精度的同时降低了网络复杂度.研究者们进一步对CNN优化,提出全卷积神经网络(fully convolutional networks,FCN).Romaguera等[12]通过FCN对输入与标签进行训练,逐像素分类以分割左心肌,十折交叉验证与RMSProp优化使网络更好地提取特征,提高了分割精度.Zhao等[13]基于FCN提出PSPNet算法,聚合了基于不同区域的上下文信息,提高挖掘全局上下文信息的能力.Badrinarayanan等[14]基于FCN提出SegNet网络,其采用对称编码译码结构来获取多尺度信息,计算效率高、分割精度更高.Ronneberger等[15]对FCN进行改进扩展,提出了U-net网络用于生物医学图像分割,医学图像数据较难获取,图像边界模糊,而U-net网络训练数据需求量小,且结合了高分辨率和低分辨率信息,分别解决了像素定位和分类问题,能更好地分割医学图像.很多研究者均采用U-net分割左心肌,并以U-net为基础进行改良.如Zhou等[16]提出Unet++,在其架构中嵌入了不同深度的U-net以减轻未知的网络深度,并且重新设计跳跃连接以在解码器子网络上聚集不同语义尺度的特征,有效减少编码译码器之间的语义鸿沟.Zottic等[17]提出GridNet,通过一个多分辨率的卷积-反卷积网格架构来学习特征,从而实现心肌分割,该网络的参数仅为原U-net的1/4,计算速度更快.Khened等提出密集U-net架构[18]、多尺度残差DenseNet网络[19]用于分割左心肌,分别改进激活、损失函数以加快网络收敛.
左心肌外部毗邻脂肪,内部常与小梁肌、乳头肌等连接,心肌特征容易被其它组织特征掩盖;此外,短轴Cine-CMR图像中心肌在底部、中部和顶部的形状差异较大,给特征提取带来困难.上述提及的方法虽然训练网络较快,但提取左心肌特征,尤其是顶部切片时,很容易连同其他组织特征一起提取,分割效果不好.针对以上难点,本文提出了一种新的压缩激励残差U形网络(squeeze-and-excitation residual U-shaped network,SERU-net)用来分割左心肌,该网络融合了压缩激励(squeeze-and-excitation,SE)模块[20]和残差模块[21],改进了U-net网络中的卷积层.SE模块使网络在提取特征时能够通过学习自动获取每个特征通道的重要程度,以提升对左心肌分割有用的特征并抑制用处不大的特征,更好地提取左心肌有效特征;残差模块有效抑制了梯度消失和训练过拟合问题,使得信息前后向传播更加顺畅,网络学习效果更好,分割精度更高.基于本文网络有助于开展后续的心肌追踪研究工作[22],而本文详细验证了该网络的有效性.
1 理论方法
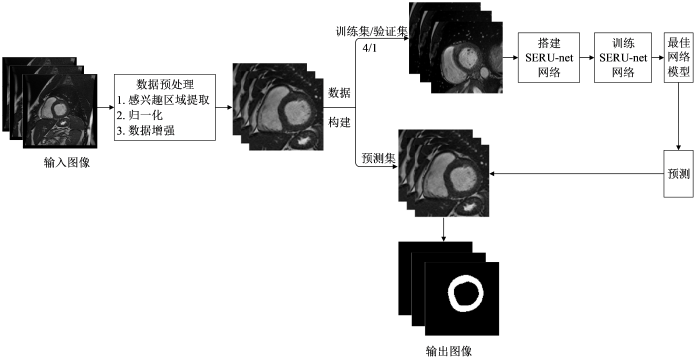
本文提出了一个结合SE模块和残差模块的深度学习网络模型,用于实现Cine-CMR图像中左心肌的自动分割.该方法的流程如图1所示,包含以下步骤:1)对原始Cine-CMR图像数据进行感兴趣区域(region of interest,ROI)提取、归一化、数据增强等预处理;2)搭建SERU-net网络,训练网络得到最佳模型;3)利用最佳模型预测得到分割结果.
图1
1.1 数据预处理
Cine-CMR图像包含左心室的同时还包含其他器官组织,且左心肌与周围组织的灰度对比度低,这将影响左心肌分割的精确度.因此,本文首先对采集的原始Cine-CMR图像进行ROI提取.首先采用霍夫圆变换确定LV中心位置,然后截取包含LV的部分图像为ROI,截取后的图像像素值大小为128×128,可以减少其他组织对后续网络训练的影响.
Cine-CMR图像数据可能存在像素灰度分布差异,给后续数据处理和网络训练带来干扰.为平衡图像像素分布,本文采用最大最小值法对ROI图像进行归一化处理,将图像的像素值取值范围从[0,255]转换为[0,1],从而加快网络训练收敛的速度.最大、最小值归一化方法的公式如下:
其中,
另外,为了抑制训练样本不足带来的问题,本文采用翻转、旋转、缩放、平移等方法对已获得的图像进行数据扩增,增加训练样本的数据量,丰富训练样本的多样性,以提高网络的泛化能力.最后,采用留出法将数据集划分为训练集、验证集、测试集.
1.2 SERU-net网络结构
U-net由FCN改进和扩展而来,由于其所需训练集数量少、对模糊边界分割效果好,被广泛用于医学图像分割.本文基于传统的U-net,融合SE模块和残差模块提出一种新的SERU-net网络,能提升对左心肌分割有用的特征提取并抑制用处不大的特征提取,从而更好地提取有效特征,进而提高分割精度.
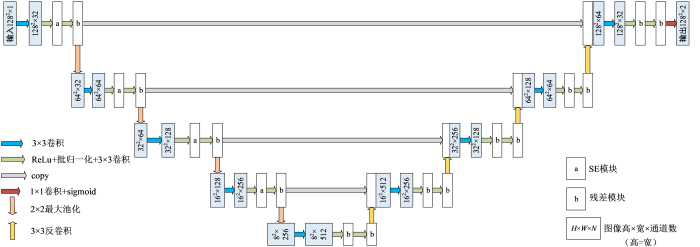
SERU-net网络的整体结构如图2所示,左侧为编码部分,右侧为解码部分.其中,a矩形框代表SE模块,输出经学习得到的不同权重调整后的特征图;b矩形框代表残差模块,本文选用由两个3×3大小的卷积层和一个短连接构成的残差.编码部分对残差模块输出的特征图采用2×2最大池化进行下采样,提取不同特征;解码部分采用3×3反卷积进行上采样,同时叠加通过跳跃连接获得的特征,融合了深层特征与浅层特征.最后通过1×1卷积和sigmoid激活函数,输出分割左心肌的概率图.
图2
图2
压缩激励残差U形网络(SERU-net)的结构
Fig. 2
Structure of squeeze-and-excitation residual U-shaped network (SERU-net)
传统的U-net网络包括基本卷积层(包括卷积核和修正线性单元ReLu)、池化层、反卷积层、以及跳跃连接.其中,卷积层用于特征提取,最大池化卷积用于特征降维,反卷积层用于还原特征图尺寸,跳跃连接用于融合浅层、深层特征.但是,左心肌外部常常毗邻脂肪,内部又常常与小梁肌、乳头肌等连接,心肌特征很容易被其它组织特征掩盖;此外,短轴Cine-CMRI图像中心肌在底部、中部和顶部的形状差异较大,给卷积层有效提取特征带来困难.针对心肌分割中的难点,考虑到SE模块有助于提取对左心肌分割有用的特征并抑制用处不大的特征,本文在普通卷积层后引入了SE模块,结构如图3(a)所示.不同于从空间维度上来提升网络性能的结构,SE模块从特征通道之间的关系入手,采用了一种“特征重标定”策略.输入
图3
图3
(a) SE模块和(b)残差模块的结构
Fig. 3
Structure of squeeze-and-excitation module (a) and residual module (b)
增加网络深度能提取到更深层次的特征,提高网络分割效果,但可能会导致网络梯度消失或训练过拟合.为了抑制网络深度增加出现的梯度消失现象,本文网络引入了残差模块,结构如图3(b)所示.假定某段神经网络的输入是x,期望输出是
为了训练SERU-net网络,采用Dice系数作为损失函数以评估图像分割值与金标准间的相似性程度.同时,利用验证集数据调整训练得到的网络的超参数,重复训练、验证,当验证集损失函数值最小时保存其网络参数,得到最佳网络模型.
将测试集输入训练好的最佳网络中,SERU-net网络输出结果为每一个像素点属于左心肌的概率,得到与输入图像同样大小的预测图.对概率图进行二值化处理,设置阈值0.5,对每个像素点进行分类得到最后的二值分割图.
2 实验部分
2.1 实验数据
本次用于左心肌分割研究的不同时相不同切片的短轴Cine-CMR图像数据均来源于康奈尔大学威尔医学院(Weill Cornell Medical College),数据的使用得到了康奈尔大学威尔医学院伦理委员会的准许,符合伦理要求.这些数据均使用GE1.5T磁共振扫描仪采集,成像序列为SSFP序列.实验数据一共包括93例病人,其中男性58例,女性35例,年龄跨度为23~93岁.具体成像参数为:图像大小256×256,层厚6~8 mm,层间距2~4 mm,每例数据包含6~10层,每层20~28个时相.采用留出法进行数据划分,75例病人数据用于训练集和验证集训练网络,其中训练集和验证集的比例为4:1,18例病人数据用于测试集分割及评价模型.
2.2 评估指标
本文以专家手动勾画结果为金标准,通过计算算法分割得到的几何指标和临床指标与金标准的差异性和相关性,对本文所提方法的分割性能进行评估.
几何指标包括:舒张末期(end diastolic,ED)与收缩末期(end systolic,ES)的Dice系数(dice metric,DM)与豪斯多夫距离(Hausdorff distance,HD),DM衡量预测结果与金标准的相似度,表示为:
|A|和|B|分别表示算法自动分割和专家手动勾画的左心肌区域,
A与B分别为预测结果与专家手动勾画的金标准,a与b分别为A与B的点,
临床指标包括:舒张末期左心室心肌质量(end diastolic-left ventricular mass,ED-LVM)和收缩末期左心室心肌质量(end systolic-left ventricular mass,ES-LVM),心肌质量为心肌容积与心肌密度(1.05 g/dL)的积,心肌容积为各层面心肌面积和图像层间距之积的总和.左心肌分割即提取出LV内膜与LV外膜,LV内膜的提取即分割LV,因此分割出左心肌的同时也完成了LV的分割.根据本文算法分割结果也能够得到LV分割结果,计算出舒张末期容积(end diastolic volume,EDV)、收缩末期容积(end systolic volume,ESV)、每搏输出量(stroke volume,SV)、射血分数(ejection fraction,EF).对算法自动分割得到的心功能参数与金标准展开相关性、一致性分析,其中一致性通过Bland-Altman图展示.
3 实验结果与讨论
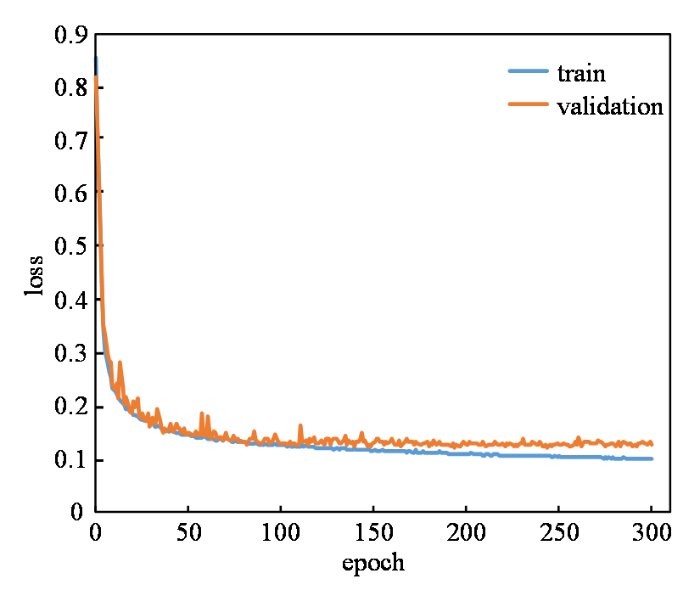
本文网络采用Dice损失函数训练300轮后得到如图4所示的损失曲线,损失值在训练开始阶段大幅下降,说明学习率合适且有助于梯度下降过程.网络学习到一定阶段后,损失曲线趋于平稳,且验证集损失曲线与训练集损失曲线几近重合,说明网络拟合较好,未出现欠拟合或过拟合现象.
图4
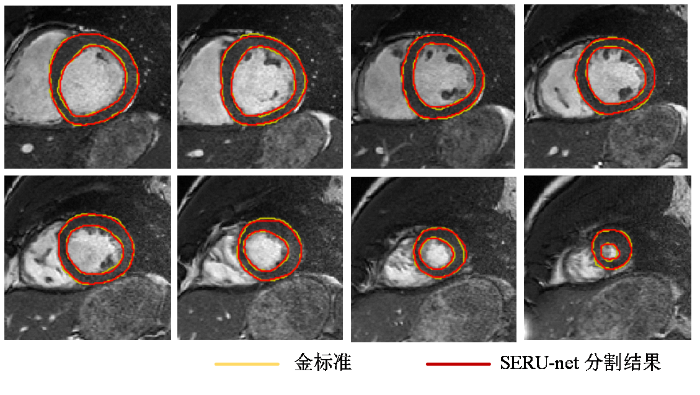
使用训练好的网络对测试集18例病人数据进行测试,并和金标准比较.从分割结果可以看出,本文算法能够依据数据学习,较精确地自动分割出左心肌,在测试集大多数图像中获得了较好的分割结果,在LV边界变得模糊且难以辨别导致分割困难的基底和顶端切片,仍获得了较好的分割结果.其中一例病例从基底到顶端的分割结果如图5所示,黄色为专家勾画的金标准,红色为本文方法分割的结果.
图5
图5
一例病例自基底至顶端的左心肌分割结果
Fig. 5
Left myocardium segmentation for a typical Cine-CMRI case from base to apex
表1 使用本文方法与其他方法得到的DM和HD的对比
Table 1
| 左心肌 | LV | ||||
|---|---|---|---|---|---|
| DM | HD/mm | DM | HD/mm | ||
| FCN[23] | 0.878 (0.031) | 3.086 (1.129) | 0.896 (0.056) | 2.920 (0.899) | |
| U-net[15] | 0.903 (0.021) | 2.736 (0.841) | 0.916 (0.050) | 2.825 (1.103) | |
| U-net++[16] | 0.894 (0.027) | 2.837 (0.617) | 0.922 (0.026) | 2.588 (0.715) | |
| SegNet[14] | 0.877 (0.034) | 3.039 (0.871) | 0.909 (0.024) | 2.974 (0.622) | |
| PSPNet[13] | 0.887 (0.025) | 2.846 (0.500) | 0.897 (0.039) | 2.846 (0.923) | |
| SERU-net(本文方法) | 0.902 (0.019) | 2.697 (0.582) | 0.928 (0.029) | 2.477 (0.796) | |
图6
图6
SERU-net分割左心肌的(a) DM、(b) HD指标箱形图
Fig. 6
Box chart of DM (a) and HD (b) indicators of left myocardium segmentation by SERU-net
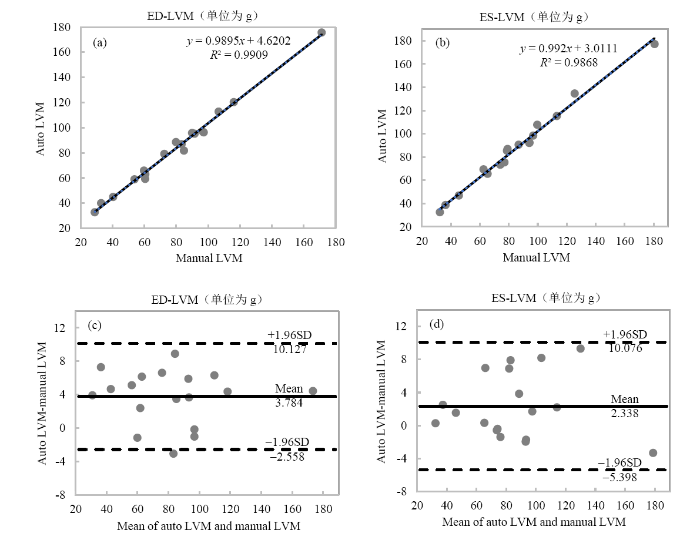
进一步地,对SERU-net算法自动分割结果与金标准进行相关性和一致性分析.图7(a)、(b)显示了使用SERU-net方法自动分割的左心室心肌质量与金标准的相关性,其中横纵坐标分别为金标准和本文算法的自动分割结果,线性回归方程和相关系数的平方R2分割在图中标出.其中,自动分割与手动分割的ED-LVM、ES-LVM的相关系数R分别为0.995、0.993,表明本文方法分割的结果与金标准具有较高的相关性.图7(c)、7(d)显示了使用SERU-net方法得到的ED-LVM、ES-LVM与金标准的一致性,其中横纵轴分别为本文算法分割结果与金标准的均值和差值,Mean表示差值的平均值,SD表示差值的标准差,Mean±1.96 SD表示“95%一致性界限”.Bland-Altman图显示,ED-LVM、ES-LVM的偏差均值分别为3.784 g、2.338 g,左心室心肌质量与金标准偏差较小,除了少数的异常值,所有测量值都在一致性界限内,表明本文方法分割的结果与金标准的一致性较好.
图7
图7
使用SERU-net算法得到的舒张末期左心室心肌质量(ED-LVM)和收缩末期左心室心肌质量(ES-LVM)与金标准的相关性和一致性分析. 相关性分析:(a) ED-LVM;(b) ES-LVM. 一致性分析:(c) ED-LVM;(d) ES-LVM
Fig. 7
Correlation analysis of (a) end diastolic-left ventricular mass, and (b) end systolic-left ventricular mass between SERU-net segmentation and ground truth. Bland-Altman analysis of (c) end diastolic-left ventricular mass, and (d) end systolic-left ventricular mass between SERU-net segmentation and ground truth
图8
图8
使用SERU-net得到的(a)舒张末期容积(EDV)、(b)收缩末期容积(ESV)、(c)每搏输出量(SV)、(d)射血分数(EF)与金标准的相关性分析
Fig. 8
Correlation analysis of (a) end diastolic volume (EDV), (b) end systolic volume (ESV), (c) stroke volume (SV), (d) ejection fraction (EF) between SERU-net segmentation and ground truth
图9
图9
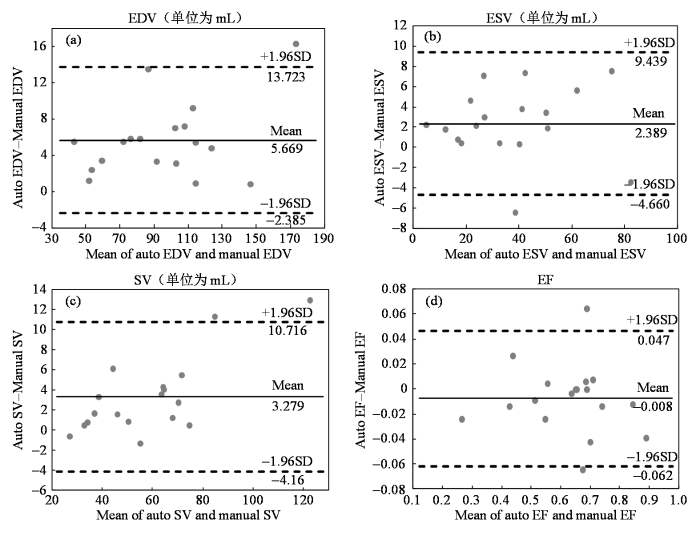
使用SERU-net得到的(a)舒张末期容积(EDV)、(b)收缩末期容积(ESV)、(c)每搏输出量(SV)、(d)射血分数(EF)与金标准的一致性分析
Fig. 9
Bland-Altman analysis of (a) end diastolic volume (EDV), (b) end systolic volume (ESV), (c) stroke volume (SV), (d) ejection fraction (EF) between SERU-net segmentation and ground truth
本文算法在测试集中的分割效果较好,但在图像质量较差的情况下(如存在部分容积效应时),网络训练时不能准确提取心肌特征,从而导致分割时也会出现误差.由于深度学习是基于海量数据的方法,学习数据越多,得到的结果越好.而医学数据量小且难获取,本文所采用的数据集较小,若能使用更大的包含不同病理的数据集训练网络,分割结果会更好,模型会更具有泛化性.
我们进一步将SERU-net的分割结果与目前主流和先进的基于深度学习神经网络的左心肌分割方法(包括FCN、U-net、U-net++、SegNet、PSPNet等)进行了比较.图10展示了各方法的分割结果,可以看出本文方法的分割结果优于其它5种方法.
图10
图10
不同方法分割同一例病例的自基底至顶端的左心肌的结果
Fig. 10
Left myocardium segmentation results of the same patient by different methods from base to apex
图11
图11
使用本文方法与其它方法分割左心肌的(a) DM、(b) HD的箱形图
Fig. 11
Box chart of DM (a) and HD (b) indicators of left myocardium segmentation using the method proposed in this paper and other methods
表2 各方法分割结果的ED_LVM、ES_LVM与金标准的相关系数(R)及偏差均值(MD)
Table 2
| Method | ED_LVM_R | ED_LVM_MD/g | ES_LVM_R | ES_LVM_MD/g |
|---|---|---|---|---|
| FCN | 0.992 | 5.793 | 0.990 | 3.854 |
| U-net | 0.993 | 4.946 | 0.990 | 3.337 |
| U-net++ | 0.993 | 4.272 | 0.991 | 2.703 |
| SegNet | 0.994 | 4.165 | 0.992 | 2.857 |
| PSPNet | 0.992 | 4.364 | 0.991 | 2.794 |
| SERU-net(本文方法) | 0.995 | 3.784 | 0.993 | 2.338 |
表3 使用各自动分割方法得到的EDV、ESV、EF与金标准的相关系数(R)及偏差均值(MD)
Table 3
| 方法 | EDV_R | EDV_MD/mL | ESV_R | ESV_MD/mL | EF_R | EF_MD |
|---|---|---|---|---|---|---|
| FCN | 0.991 | 5.728 | 0.973 | 4.407 | 0.980 | -0.023 |
| U-net | 0.995 | 8.333 | 0.980 | 6.897 | 0.995 | -0.026 |
| U-net++ | 0.995 | 7.390 | 0.978 | 4.428 | 0.969 | -0.021 |
| SegNet | 0.993 | 9.360 | 0.991 | 5.644 | 0.991 | -0.023 |
| PSPNet | 0.990 | 6.780 | 0.986 | 5.449 | 0.977 | -0.030 |
| SERU-net(本文方法) | 0.994 | 5.669 | 0.986 | 2.389 | 0.983 | -0.008 |
4 结论
本文提出了一种新的压缩激励残差U-net网络结构,用于短轴Cine-CMRI图像的心肌分割.该网络将 U-net作为主干,融入了SE模块,自动学习特征重要性并对原始特征进行权重分配,从特征通道维度上提升了网络性能.同时,结合残差模块解决了网络反向传播中的梯度弥散问题,有效抑制了网络退化.结果证明,该网络分割结果与金标准之间具备较高一致性,成功验证了算法的可行性和准确性.与其他方法比较,我们提出的方法能够快速准确地分割出左心肌,且具有更高的准确性和鲁棒性.为了进一步提高分割的准确性,今后的工作可按以下两项进行:一是使用更大的包含不同病理的数据集训练网络,提高模型泛化性能;二是进一步优化网络结构,更好地提取图像特征.
利益冲突
无
参考文献
Summary of the 2018 report on cardiovascular diseases in China
[J].
《中国心血管病报告2018》概要
[J].
Accelerated cardiac CINE imaging with CAIPIRINHA and partial parallel acquisition
[J].
基于同时多层激发和并行成像的心脏磁共振电影成像
[J].
Progress of right ventricle segmentation from short-axis images acquired with cardiac cine MRI
[J].
基于心脏磁共振短轴电影图像的右心室分割新进展
[J].
A review of segmentation methods in short axis cardiac MR images
[J].
DOI:10.1016/j.media.2010.12.004
PMID:21216179
[本文引用: 1]

For the last 15 years, Magnetic Resonance Imaging (MRI) has become a reference examination for cardiac morphology, function and perfusion in humans. Yet, due to the characteristics of cardiac MR images and to the great variability of the images among patients, the problem of heart cavities segmentation in MRI is still open. This paper is a review of fully and semi-automated methods performing segmentation in short axis images using a cardiac cine MRI sequence. Medical background and specific segmentation difficulties associated to these images are presented. For this particularly complex segmentation task, prior knowledge is required. We thus propose an original categorization for cardiac segmentation methods, with a special emphasis on what level of external information is required (weak or strong) and how it is used to constrain segmentation. After reviewing method principles and analyzing segmentation results, we conclude with a discussion and future trends in this field regarding methodological and medical issues.Copyright © 2010 Elsevier B.V. All rights reserved.
Fast segmentation of the left ventricle in cardiac MRI using dynamic programming
[J].DOI:10.1016/j.cmpb.2017.10.028 URL [本文引用: 1]
Left ventricle segmentation via two-layer level sets with circular shape constraint
[J].
DOI:S0730-725X(17)30011-5
PMID:28108373
[本文引用: 1]

This paper proposes a circular shape constraint and a novel two-layer level set method for the segmentation of the left ventricle (LV) from short-axis magnetic resonance images without training any shape models. Since the shape of LV throughout the apex-base axis is close to a ring shape, we propose a circle fitting term in the level set framework to detect the endocardium. The circle fitting term imposes a penalty on the evolving contour from its fitting circle, and thereby handles quite well with issues in LV segmentation, especially the presence of outflow track in basal slices and the intensity overlap between TPM and the myocardium. To extract the whole myocardium, the circle fitting term is incorporated into two-layer level set method. The endocardium and epicardium are respectively represented by two specified level contours of the level set function, which are evolved by an edge-based and a region-based active contour model. The proposed method has been quantitatively validated on the public data set from MICCAI 2009 challenge on the LV segmentation. Experimental results and comparisons with state-of-the-art demonstrate the accuracy and robustness of our method.Copyright © 2017 Elsevier Inc. All rights reserved.
Automatic left ventricle segmentation using iterative thresholding and an active contour model with adaptation on short-axis cardiac MRI
[J].DOI:10.1109/TBME.2009.2014545 URL [本文引用: 1]
Deep learning techniques for automatic MRI cardiac multi-structures segmentation and diagnosis: Is the problem solved?
[J].DOI:10.1109/TMI.2018.2837502 URL [本文引用: 1]
A combined deep-learning and deformable-model approach to fully automatic segmentation of the left ventricle in cardiac MRI
[J].
DOI:S1361-8415(16)00012-8
PMID:26917105
[本文引用: 1]

Segmentation of the left ventricle (LV) from cardiac magnetic resonance imaging (MRI) datasets is an essential step for calculation of clinical indices such as ventricular volume and ejection fraction. In this work, we employ deep learning algorithms combined with deformable models to develop and evaluate a fully automatic LV segmentation tool from short-axis cardiac MRI datasets. The method employs deep learning algorithms to learn the segmentation task from the ground true data. Convolutional networks are employed to automatically detect the LV chamber in MRI dataset. Stacked autoencoders are used to infer the LV shape. The inferred shape is incorporated into deformable models to improve the accuracy and robustness of the segmentation. We validated our method using 45 cardiac MR datasets from the MICCAI 2009 LV segmentation challenge and showed that it outperforms the state-of-the art methods. Excellent agreement with the ground truth was achieved. Validation metrics, percentage of good contours, Dice metric, average perpendicular distance and conformity, were computed as 96.69%, 0.94, 1.81 mm and 0.86, versus those of 79.2-95.62%, 0.87-0.9, 1.76-2.97 mm and 0.67-0.78, obtained by other methods, respectively.Copyright © 2016 Elsevier B.V. All rights reserved.
Combining deep learning and level set for the automated segmentation of the left ventricle of the heart from cardiac cine magnetic resonance
[J].
DOI:S1361-8415(16)30038-X
PMID:27423113
[本文引用: 1]

We introduce a new methodology that combines deep learning and level set for the automated segmentation of the left ventricle of the heart from cardiac cine magnetic resonance (MR) data. This combination is relevant for segmentation problems, where the visual object of interest presents large shape and appearance variations, but the annotated training set is small, which is the case for various medical image analysis applications, including the one considered in this paper. In particular, level set methods are based on shape and appearance terms that use small training sets, but present limitations for modelling the visual object variations. Deep learning methods can model such variations using relatively small amounts of annotated training, but they often need to be regularised to produce good generalisation. Therefore, the combination of these methods brings together the advantages of both approaches, producing a methodology that needs small training sets and produces accurate segmentation results. We test our methodology on the MICCAI 2009 left ventricle segmentation challenge database (containing 15 sequences for training, 15 for validation and 15 for testing), where our approach achieves the most accurate results in the semi-automated problem and state-of-the-art results for the fully automated challenge.Crown Copyright © 2016. Published by Elsevier B.V. All rights reserved.
Deep fusion net for multi-atlas segmentation: Application to cardiac MR images
[C]//
Myocardial segmentation in cardiac magnetic resonance images using fully convolutional neural networks
[J].DOI:10.1016/j.bspc.2018.04.008 URL [本文引用: 1]
Pyramid scene parsing network
[J].
SegNet: A deep convolutional encoder-decoder architecture for image segmentation
[J].
DOI:10.1109/TPAMI.2016.2644615
PMID:28060704
[本文引用: 2]

We present a novel and practical deep fully convolutional neural network architecture for semantic pixel-wise segmentation termed SegNet. This core trainable segmentation engine consists of an encoder network, a corresponding decoder network followed by a pixel-wise classification layer. The architecture of the encoder network is topologically identical to the 13 convolutional layers in the VGG16 network [1]. The role of the decoder network is to map the low resolution encoder feature maps to full input resolution feature maps for pixel-wise classification. The novelty of SegNet lies is in the manner in which the decoder upsamples its lower resolution input feature map(s). Specifically, the decoder uses pooling indices computed in the max-pooling step of the corresponding encoder to perform non-linear upsampling. This eliminates the need for learning to upsample. The upsampled maps are sparse and are then convolved with trainable filters to produce dense feature maps. We compare our proposed architecture with the widely adopted FCN [2] and also with the well known DeepLab-LargeFOV [3], DeconvNet [4] architectures. This comparison reveals the memory versus accuracy trade-off involved in achieving good segmentation performance. SegNet was primarily motivated by scene understanding applications. Hence, it is designed to be efficient both in terms of memory and computational time during inference. It is also significantly smaller in the number of trainable parameters than other competing architectures and can be trained end-to-end using stochastic gradient descent. We also performed a controlled benchmark of SegNet and other architectures on both road scenes and SUN RGB-D indoor scene segmentation tasks. These quantitative assessments show that SegNet provides good performance with competitive inference time and most efficient inference memory-wise as compared to other architectures. We also provide a Caffe implementation of SegNet and a web demo at http://mi.eng.cam.ac.uk/projects/segnet.
U-Net: Convolutional networks for biomedical image segmentation
[C]//
UNet++: A nested U-net architecture for medical image segmentation
[C]// Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support:4th International Workshop.
GridNet with automatic shape prior registration for automatic MRI cardiac segmentation
[C]//
Densely connected fully convolutional network for short-axis cardiac cine MR image segmentation and heart diagnosis using random forest
[C]//
Fully convolutional multi-scale residual DenseNets for cardiac segmentation and automated cardiac diagnosis using ensemble of classifiers
[J].
DOI:S1361-8415(18)30848-X
PMID:30390512
[本文引用: 1]

Deep fully convolutional neural network (FCN) based architectures have shown great potential in medical image segmentation. However, such architectures usually have millions of parameters and inadequate number of training samples leading to over-fitting and poor generalization. In this paper, we present a novel DenseNet based FCN architecture for cardiac segmentation which is parameter and memory efficient. We propose a novel up-sampling path which incorporates long skip and short-cut connections to overcome the feature map explosion in conventional FCN based architectures. In order to process the input images at multiple scales and view points simultaneously, we propose to incorporate Inception module's parallel structures. We propose a novel dual loss function whose weighting scheme allows to combine advantages of cross-entropy and Dice loss leading to qualitative improvements in segmentation. We demonstrate computational efficacy of incorporating conventional computer vision techniques for region of interest detection in an end-to-end deep learning based segmentation framework. From the segmentation maps we extract clinically relevant cardiac parameters and hand-craft features which reflect the clinical diagnostic analysis and train an ensemble system for cardiac disease classification. We validate our proposed network architecture on three publicly available datasets, namely: (i) Automated Cardiac Diagnosis Challenge (ACDC-2017), (ii) Left Ventricular segmentation challenge (LV-2011), (iii) 2015 Kaggle Data Science Bowl cardiac challenge data. Our approach in ACDC-2017 challenge stood second place for segmentation and first place in automated cardiac disease diagnosis tasks with an accuracy of 100% on a limited testing set (n=50). In the LV-2011 challenge our approach attained 0.74 Jaccard index, which is so far the highest published result in fully automated algorithms. In the Kaggle challenge our approach for LV volume gave a Continuous Ranked Probability Score (CRPS) of 0.0127, which would have placed us tenth in the original challenge. Our approach combined both cardiac segmentation and disease diagnosis into a fully automated framework which is computationally efficient and hence has the potential to be incorporated in computer-aided diagnosis (CAD) tools for clinical application.Copyright © 2018 Elsevier B.V. All rights reserved.
Squeeze-and-excitation networks
[J].
Deep residual learning for image recognition
[C]//
Motion tracking of left myocardium in cardiac cine magnetic resonance image based on displacement flow U-Net and variational autoencoder
[J].DOI:10.7498/aps URL [本文引用: 1]
基于位移流U-Net和变分自动编码器的心脏电影磁共振图像左心肌运动追踪
[J].DOI:10.7498/aps URL [本文引用: 1]
Fully convolutional networks for semantic segmentation
[J].DOI:10.1109/TPAMI.2016.2572683 URL [本文引用: 1]