引言
当前膝关节局部SAR估计的主要方法是通过构建特异性的膝关节模型并以此进行电磁仿真[7],这就需要基于膝关节MR图像进行组织分割以重建模型. 一些情况下可以根据获得的低场MR图像重建模型,以此进行高场扫描前的电磁仿真与局部SAR估计. 低场图像组织对比度较低,且由于膝关节结构复杂,一些组织在图像上模糊且间断,难以准确分割. 组织简化可以适应信噪比较低的图像,减少人工标注的误差与时间消耗.学术界经常采用组织简化策略来进行局部SAR估计[8,9].Carluccio等[10]使用只有肌肉、脂肪和肺的简化模型,与原始模型相比得出了较接近的局部SAR估计[11].表明当用肌肉替换除骨骼、脂肪和肺以外的所有组织类型时,人体躯干模型中的局部SAR值无显著差异. 对于膝关节,考虑到组织的介电特性[12],可以简化为肌肉、脂肪和骨骼三种有代表性的组织,这适用于图像信噪比较低的情况.
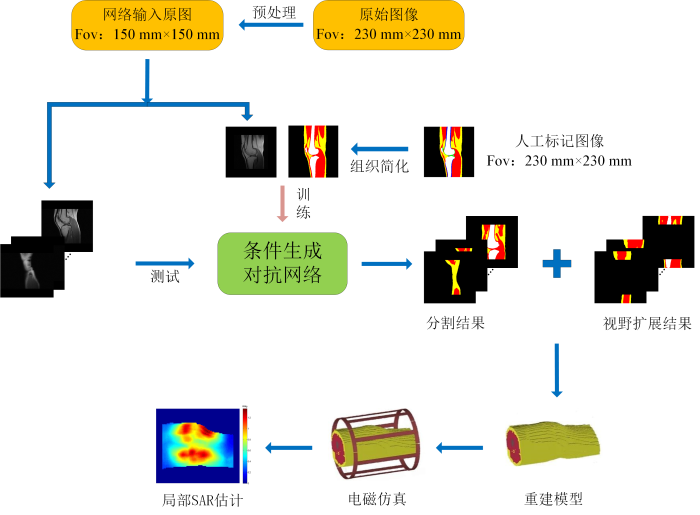
我们前期基于230 mm视野图像进行了图像分割与局部SAR估计的研究[23]. 为了提高小视野图像构建模型的局部SAR估计的准确度,本文针对常用的150 mm视野图像,提出了一种基于条件生成对抗网络的图像分割和视野扩展方法,该方法采用“肌肉-脂肪-骨骼”组织简化,集成了融合注意力机制的U型网络(U-Net)为生成网络,将对抗损失和绝对值损失函数(L1损失)结合作为目标损失函数,对输入图像进行语义分割,并在图像两端生成组织扩展到230 mm视野,从而构建更长的模型.实验中,分别对所提方法和三种对比方法构建的膝关节模型进行了电磁仿真和局部SAR估计.
1 方法
1.1 数据集
本研究使用的膝关节数据集与之前的研究[23]采用的数据集相同,为矢位纵向弛豫(T1)加权自旋回波图像,来自于67位志愿者,志愿者的平均年龄为46.3岁. 选择数据集中48位志愿者的图像作为训练集,19位志愿者的图像作为测试集. 图像的扫描于2019年在张家口仁爱医院的0.35 T MRI系统上进行,获得了院方许可,实验之前均对每一位志愿者进行了告知,并获得了他们的同意,实验中由影像科医生负责扫描,以确保安全.
扫描图像的视野为230 mm×230 mm,层厚为4 mm,每位志愿者的图像切片数为30,图像尺寸为384×384,体素尺寸为0.6 mm×0.6 mm×4 mm.在本研究中,电磁仿真所采用的鸟笼线圈长度为180 mm,这样基于230 mm视野所构建模型的局部SAR基本上接近整个人腿的情况,可以作为对比的基准.实际扫描时的图像视野常为150 mm×150 mm,对于230 mm而言属于视野较小的情况,因此这里研究从150 mm视野扩展到230 mm视野.在网络训练与测试中,根据视野扩展的需求将原始数据集的视野修剪为150 mm×150 mm.
所有的图像由专业影像人员标注为肌肉、脂肪、半月板、软骨、皮质骨和松质骨六种组织,在此基础上进行组织简化,将骨骼与脂肪之外的组织全部归为肌肉,因此得到肌肉、脂肪和骨骼三种组织的标注图像.
1.2 所提方法的总体结构
所提膝关节模型构建方法的整体流程图如图1所示,训练集中视野为150 mm×150 mm的原始图像与其对应的视野为230 mm×230 mm的人工标注“肌肉-脂肪-骨骼”图像作为输入数据,以训练条件生成对抗网络. 该网络对三种组织进行分割的同时,在图像头-足方向的两端生成包含三种组织的区域,从而扩展视野. 在测试时,每一位志愿者的所有图像输入网络,得到视野为230 mm×230 mm的“肌肉-脂肪-骨骼”分类图像,将每一位志愿者的所有分类图像切片合在一起,得到长度为230 mm而非150 mm的膝关节模型,将膝关节模型放置在射频发射线圈模型中,进行电磁仿真并计算局部SAR分布.
图1
图1
所提方法的膝关节模型构建整体流程图
Fig. 1
The whole flowchart of knee joint model construction using the proposed method
1.3 条件生成对抗网络(CGAN)
CGAN由生成网络G和判别网络D两个子网络组成,生成网络G负责从噪声中生成具有先验信息的合成样本,判别网络D是一个分类器,负责判别传入网络的数据是真实数据还是合成样本.在训练过程中,两个子网络同时训练但目标相反.CGAN的优化目标如(1)式 [22]所示:
其中x表示真实数据,y表示条件,z表示随机噪声,
本文使用的CGAN的结构如图2所示. 生成网络以原始小视野图像(Fov为150 mm×150 mm)作为条件y,从噪声z中合成原始MR图像的分割扩展图像
图2
图2
所提出的CGAN的结构. 根据判别网络输出的0/1矩阵计算对抗损失,误差反向传播给生成网络和判别网络,使用人工标注图像和生成网络生成结果计算L1损失,反向传播给生成网络,根据梯度下降原理不断调整网络参数
Fig. 2
The architecture of the proposed CGAN. The countermeasure loss is calculated according to the 0/1 matrix of the output of the discriminator network. The error is propagated back to the generator network and the discriminator network, and the L1 loss is calculated by using the artificially labeled image and the generating result of the generator network, transmitting back to the generator network, and the network parameters are continuously adjusted according to the gradient descent principle
1.4 生成网络
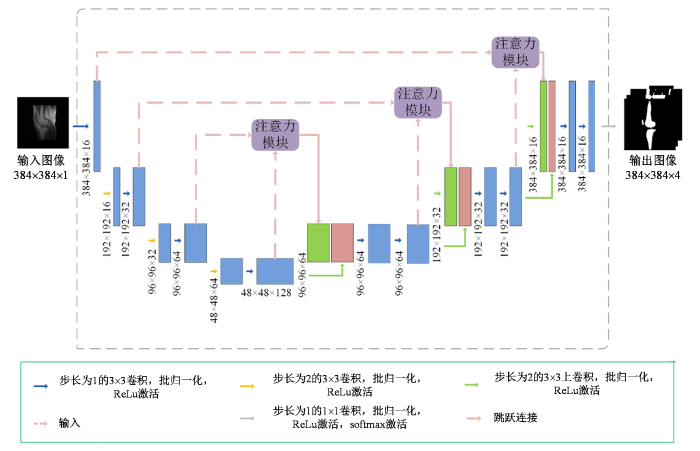
如图3所示,本文使用的生成网络为一个4层的U-Net,采用原始小视野MR图像作为条件y输入,而输入的随机噪声z体现在网络训练与测试的dropout步骤中. 每层进行两次卷积操作,使用边缘0填充的卷积方式,可保持网络的输入图像与输出图像大小一致. 使用步长为2的卷积操作代替池化操作进行下采样,可以避免边缘特征在传播中逐渐消失. 每次卷积操作后都进行批归一化和线性整流激活(ReLu激活).在上采样过程中每个跳跃连接末端加入注意力模块,较浅层的特征图监督较深层的特征图,将权重主要分配在待分割的区域,减小背景的权重来优化分割. 最后一个解码器层的输出附加了softmax激活函数,该函数为每个输出标签生成输出概率图.
图3
图3
生成网络的结构. 网络的输入为原始灰度图像,输出肌肉、脂肪、骨骼与背景4种图像
Fig. 3
The structure of generator network. The network input is the original gray-scale image, and the output consists of muscle, fat, bone and background images
1.5 注意力模块
在电磁仿真中,如果背景区域被错误分类为某种膝关节组织,会导致SAR估计产生明显的错误.为此,在网络设计中引入注意力模块[24],可以将注意力集中在特定的区域,抑制背景区域的特征激活.注意力模块的结构如图4所示.使用上一层编码部分的输出特征图el-1作为门控信号,解码部分特征图dl进行上采样获得同样的尺寸,对它们进行卷积操作从而实现线性变换,然后使用ReLu函数激活,经过卷积层后采用sigmoid激活函数能够在训练过程中使参数更好的收敛,输出注意力系数矩阵α,使其与dl相乘,能够确定感兴趣区域.在U-Net的跳跃连接中加入注意力模块,从浅层次的特征图提取上下文信息,并突出显示感兴趣区域的位置,随后通过跳跃连接将以多个比例提取的特征图合并,可以实现更精确的分割.
图4
图4
生成网络中使用的注意力模块. 根据解码部分特征图dl与其上一层编码部分特征图el-1作为输入,计算注意力系数矩阵α,el-1与注意力系数矩阵α逐像素相乘来选择聚焦区域
Fig. 4
The attention modules used in generator network. The attention coefficient matrix α is calculated by using the decoded partial characteristic graph dl and the coded partial characteristic graph el-1 as the input, el-1 multiplies the attention coefficient matrix α pixel by pixel to select the focus region
1.6 判别网络
判别网络采用了patch-GAN的思想,为5层的卷积神经网络结构,采用小视野原始MR图像与大视野人工标注图像的组合,或其与生成网络生成的分割扩展图像(Fov为230 mm×230 mm)的组合作为输入,使用步长为2的卷积实现每层的下采样,对每次卷积后的结果均进行了ReLu激活与批归一化,最后一层为一个卷积层,其输出后接sigmoid函数,最终输出为一个由0与1组成的二维矩阵,矩阵中的每一个数字都对应了输入图像组合的一个区域的判别结果,矩阵越大,对应的区域越小,这样训练可以使模型更关注图像的细节. 判别网络的网络结构如图5所示.
图5
1.7 损失函数的设计
在训练中要解决(1)式的极大极小问题:固定生成网络,通过合成和人工标注的分割扩展图像训练判别网络,使判别网络的损失函数最大化;或者固定判别网络,通过合成分割扩展图像训练生成网络,使生成网络的损失函数最小化,这两个损失为对抗损失.
除对抗损失外,在损失函数中添加L1损失,可以帮助生成器捕捉上下文信息,这有利于补充图像中头-足方向需要生成的扩展区域.损失函数的定义如(2)式所示:
上式中λ为L1损失的权重系数.
2 实验
2.1 网络训练
使用Python语言编写网络模型,在TensorFlow平台上训练和预测. 用于实验的计算机配置有4核英特尔i7-4 770处理器和具有16 GB内存的GeForce GTX显卡. 生成网络包含4个编码层和解码层,在跳跃连接中加入了注意力模块,卷积核大小为3×3. 判别网络为5层的卷积神经网络,卷积内核大小为3×3,初始卷积核数量设置为16. 生成网络和判别网络都使用自适应矩估计优化器(ADAM优化器)训练,学习率为0.001,L1损失函数的权重λ设置为10.在训练过程中,为了使生成网络与判别网络的性能尽量均衡,我们试验了不同的生成网络训练次数与判别网络训练次数之比,最终设置为每训练3次生成网络就训练一次判别网络.
2.2 分割的评价指标
以人工标注结果作为基准,图像分割性能通过Dice系数(DCC)与真阳性率(TPR)进行评估,这两项指标计算公式如(3)和(4)式所示:
其中T是人工标注结果,U是网络的分割结果.
2.3 电磁仿真与SAR计算
将生成对抗网络的输出结果逐片组合,形成最终的膝关节模型.在电磁仿真中,采用3 T的膝关节发射线圈模型,这是一个正交鸟笼线圈,采用集总电容模式.其长度和半径分别为180 mm和87.5 mm.
构造的膝关节模型置于线圈的中心.两个1 V幅度的正交正弦波分别注入线圈的两个端口.线圈内部的电场是两个端口产生的电场的总和.
电磁仿真完成后,即可基于仿真结果计算局部SAR分布.
SAR计算公式如(5)式[25]所示:
其中σ是组织的电导率,ρ是组织的密度,
局部SAR值是SAR的平均,用SAR10g表达,是指某一体素p周边10 g组织的SAR的平均值,其计算公式如(6)式[25]所示:
其中V代表一个立方区域,该立方区域以p为中心并有10 g质量,mq代表V中任意一体素q的质量.
3 结果与讨论
3.1 分割结果
对150 mm视野区域磁共振图像的准确分割确保了所构建的模型在中心150 mm范围内与原始模型的相似度,对于局部SAR的精准估计是必要的. 为了评估所提出的架构的分割性能,对所提方法与经典的U-Net,以及融合注意力模块的U型网络Attention U-Net[24]进行比较(U-Net网络和Attention U-Net网络为4层,卷积核大小为7×7),基于原始的150 mm×150 mm视野图像测试集,计算了各方法的分割性能评价指标,如表1所示.根据表1可以看出,比起U-Net与Attention U-Net,所提方法总体上的分割性能是最佳的,肌肉与脂肪的DCC、脂肪与骨骼的TPR明显大于另外两种方法.
表1 各种方法的分割结果评价指标(所提方法、U-Net、Attention U-Net相比人工标注结果)
Table 1
| 分割方法 | 评价指标 | 肌肉 | 脂肪 | 骨骼 |
|---|---|---|---|---|
| 所提方法 | DCC | 0.8798 | 0.9135 | 0.9022 |
| TPR | 0.8915 | 0.9198 | 0.9114 | |
| U-Net | DCC | 0.8472 | 0.9040 | 0.9006 |
| TPR | 0.8841 | 0.8785 | 0.8921 | |
| Attention U-Net | DCC | 0.8578 | 0.9022 | 0.9058 |
| TPR | 0.9059 | 0.8611 | 0.8991 |
图6
图6
一位志愿者(女性,21岁)的一个膝关节MR切片基于不同方法的分割结果. 其中黄色代表脂肪,红色代表肌肉,白色代表骨骼,视野为150 mm×150 mm
Fig. 6
Segmentation results of one knee section of a female volunteer based on different methods, where yellow represents fat, red represents muscle, and white represents bone, with a field of view of 150 mm×150 mm
3.2 SAR估计结果
3.1节已经验证了本文提出的架构在分割中优于经典的U-Net和比较先进的Attention U-Net,由于本文在对膝关节磁共振图像进行分割的同时,还在头-足两端生成了扩展的部分,对于模型的合理外推可以使模型长度大于线圈长度,同时避免在模型边界处的问题,防止电流在截断处形成伪环,导致该区域的局部SAR升高[26],为了验证所提出的方法生成的更长模型对SAR估计的改进,我们将所提方法与以下三种方法做局部SAR估计的对比.
(1)方法1,使用U-Net网络将Fov为150 mm×150 mm的小视野膝关节图像分割为肌肉、脂肪和骨骼三种组织,基于网络输出结果构建150 mm长的膝关节模型,该对比的目的在于研究膝关节模型外推对局部SAR估计精度的影响.
(3)方法3,在U-Net分割的基础上采用头-足方向两端的重复外推,具体做法是:使用U-Net网络分割的视野为150 mm×150 mm的图像,沿头-足方向由两端边界图像向外重复外扩,两端分别扩展40 mm,进而构建230 mm长度的膝关节模型.采用这种方法可以保持边界上组织分布的连续性.
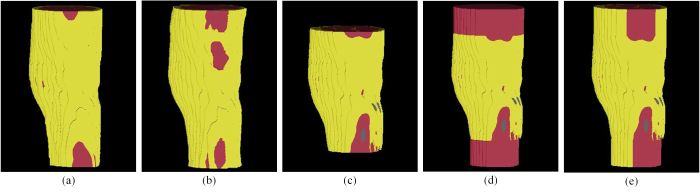
使用所提方法与对比方法得到的膝关节模型的示例如图7所示.
图7
图7
对一位志愿者(女性,21岁)使用各种不同方法构建的膝关节模型. (a)人工标注;(b)所提方法;(c)方法1;(d)方法2;(e)方法3
Fig. 7
Models of the knee joint of a female volunteer constructed with different methods. (a) artificial labeling; (b) the proposed method;(c) method 1; (d) method 2; (e) method 3
以人工标注的230 mm膝关节模型(六种组织)为基准,所提方法与对比方法的SAR10g最大值的相对误差的统计值如表2所示.
表2 最大SAR10g的相对误差的统计(所提方法和对比方法相比人工标注结果)
Table 2
| 所提方法 | 方法1 | 方法2 | 方法3 | |
|---|---|---|---|---|
| 平均值 | 0.0696 | 0.2932 | 0.0783 | 0.1066 |
| 标准差 | 0.0542 | 0.1206 | 0.0527 | 0.0525 |
方法2与方法3均为在150 mm视野区域分割的基础上进行扩展以构建模型,尽管方法3两端最后一层重复外推的结果包含了肌肉、脂肪与骨骼三种组织,但最后一层的组织分布与外推区域实际的组织分布有一定差异,这种差异对局部SAR的估计精度会造成影响.这种情况在方法2中也存在,由于局部SAR取决于体内电场分布,而导致电场分布的因素比较复杂,因此通过电磁仿真进行两个方法的比较.
表3 不同模型的最大SAR10g的统计
Table 3
| 模型 | Duke左腿 | Duke右腿 | Ella左腿 | Ella右腿 |
|---|---|---|---|---|
| 230 mm-全组织 | 1.4973 | 1.2078 | 1.0765 | 1.3145 |
| 230 mm-简化 | 1.4404 | 1.3555 | 1.1862 | 1.4363 |
| 230 mm-肌肉 | 1.4500 | 1.3261 | 1.1587 | 1.3676 |
| 230 mm-两端重复外推 | 1.5249 | 1.3940 | 1.2387 | 1.4592 |
| 150 mm-简化 | 1.9746 | 1.9019 | 1.5773 | 1.8345 |
根据表3,计算每一种模型与230mm-全组织模型的SAR10g最大值的相对误差,并且求出相对误差的平均值,可以得出150 mm-简化模型与230 mm-全组织模型的相对误差为0.438 6,说明模型长度对局部SAR有较大影响.对于不同外推方法,230 mm-肌肉和230 mm-两端重复外推模型与230 mm-全组织模型的相对误差分别为0.061 6和0.108 3,采用肌肉填充的方法误差更小,与前文的结论是一致的.此外,230 mm-简化模型与全组织模型的相对误差为0.088 7,这也证明了“肌肉-脂肪-骨骼”的组织简化方法对局部SAR没有显著影响.
我们前期的研究[23]是基于230 mm视野的原始图像分割六种组织以构建模型,得到的局部SAR最大值的相对误差的均值为0.036 6,标准差为0.035 7.与本文所提方法的偏差为0.033±0.035 7,这表明图像视野扩展与组织简化确实对局部SAR的估计精度有一定影响,这可以归因于以下事实:之前研究[23]是采用230 mm视野图像重建模型,由于所用鸟笼线圈长度为180 mm,所以不需要扩展图像.然而,实际临床膝关节扫描中大部分是采用150 mm甚至更小的视野.本文针对这种情况,采用230 mm视野图像数据集(与文献[23]相同)作为对照的基准,从中裁剪得到150 mm视野图像以模拟实际临床扫描得到的图像,再通过生成头-足各40 mm的组织得到230 mm模型,因此局部SAR预测精度会有下降.另一方面,采用“肌肉-脂肪-骨骼”的组织简化方案,必然会导致局部SAR估计结果与高度详细的模型之间有偏差,然而简化可以适应信噪比较低的图像,减少人工标注的误差与时间消耗,具有较强的实用意义.
图8
图8
同一位志愿者(女性,21岁)的膝关节SAR10g分布图.从上到下为连续5层的分布,从左到右的每一列分别为基于人工标注、所提方法、方法1、方法2和方法3的结果
Fig. 8
SAR10g distribution of the same volunteer’s knee. From top to bottom are the distribution of five consecutive MR slices, from left to right, are the results based on artificially labeling, the proposed method, method 1, method 2 and method 3 respectively
图9显示了该志愿者的各种方法所建模型的SAR10g的最大密度投影. 人工标注、所提方法、方法1、方法2以及方法3的SAR10g的最大值分别为1.390 W/kg、1.456 W/kg、1.773 W/kg、1.477 W/kg和1.522 W/kg. 所提方法与人工标注的结果最为接近,同时,热点区域的位置基本相同. 这说明了所提方法的有效性.
图9
图9
同一位志愿者(女性,21岁)的各种方法重建模型的SAR10g的最大密度投影图. (a)人工标注;(b)所提方法;(c)方法1;(d)方法2;(e)方法3
Fig. 9
The same volunteer’s maximum intensity projection of SAR10g by (a) artificially labeling; (b) the proposed method; (c) method 1; (d) method 2; (e) method 3
3.3 讨论
为了准确地估计局部SAR,快速而精准地构建个体特异性的膝关节模型具有重要意义.考虑到临床实际扫描中采集的膝关节图像视野主要在150 mm及以下,当采用180 mm长的鸟笼线圈时,基于该视野图像构建的模型的局部SAR估计误差较大.针对这种情况,要求对膝关节MR图像进行自动分割,对分割结果进行图像区域的扩展,构建更长的膝关节模型以实现更准确的局部SAR估计.
近年来,一些基于全卷积神经网络的模型如用于图像分割的深度卷积编码器-解码器架构(SegNet)、U-Net等广泛用于膝关节磁共振图像分割. 不同于这种类型的网络,生成对抗网络是由生成网络和判别网络两个网络组成,期待的结果是由生成网络学习原始数据分布得到的,判别网络在训练过程中对生成网络的结果进行判别,指导生成网络生成数据.通过对抗训练使生成网络模拟原始数据分布,可以生成更加逼真的图像. 此外视野扩展与分割采用同一生成对抗网络架构,在该网络中外推扩展与图像分割相互促进,对于图像分割任务来说,可以在生成外推部分的组织分布时更好的学习分割部分的特征图,提升磁共振图像上的分割性能.通过分割任务,网络可以更好地学习图像的语义信息和上下文关系,更好地理解图像中的结构从而帮助外推扩展更好地生成外扩的部分,提高所构建模型的外推部分的相似度.因此不仅能够在膝关节磁共振图像的组织分割中取得好的结果,也能够在小视野的膝关节图像的头-足两端生成更接近真实组织分布的扩展区域,从而构建更接近实际人腿的模型.
电磁仿真与局部SAR估计结果表明,采用“肌肉-脂肪-骨骼”组织简化,基于条件生成对抗网络进行分割与视野扩展所构建的膝关节模型,得到的SAR10g最大值与人工标注模型相对误差较小,热点区域基本相同,这说明所提出的方法在一定程度上能够得到比较接近人工标注的局部SAR估计.未来我们将扩大数据集,丰富志愿者的类型,对所提方法进一步验证与优化,得到更好的局部SAR估计结果.
4 结论
本文提出了一种基于条件生成对抗网络的全自动组织分割与视野扩展方法,用于更精准的膝关节局部SAR估计. 在将膝关节MR图像简化分类为肌肉、脂肪和骨骼三种组织的同时,还在头-足方向的两端生成包含组织的图像区域,扩大了膝关节模型的尺寸,使其更接近于实际人腿的情况. 局部SAR估计的结果表明了所提方法的有效性.
利益冲突
无
参考文献
MRI and 18F-FDG PET/CT findings of a giant cell tumor of the tendon sheath of the knee joint (pigmented villonodular synovitis): a case report and literature review
[J].
The development of ultra-high field magnetic resonance imaging
[J].
磁共振成像发展与超高场磁共振成像技术
[J].
Automatic segmentation of knee joint synovial magnetic resonance images based on 3D VNetTrans
[J].
基于3D VNetTrans的膝关节滑膜磁共振图像自动分割
[J].
Parallel transmission for ultrahigh-field imaging
[J].
DOI:10.1002/nbm.3313
PMID:25989904
[本文引用: 1]

The development of MRI systems operating at or above 7 T has provided researchers with a new window into the human body, yielding improved imaging speed, resolution and signal-to-noise ratio. In order to fully realise the potential of ultrahigh-field MRI, a range of technical hurdles must be overcome. The non-uniformity of the transmit field is one of such issues, as it leads to non-uniform images with spatially varying contrast. Parallel transmission (i.e. the use of multiple independent transmission channels) provides previously unavailable degrees of freedom that allow full spatial and temporal control of the radiofrequency (RF) fields. This review discusses the many ways in which these degrees of freedom can be used, ranging from making more uniform transmit fields to the design of subject-tailored RF pulses for both uniform excitation and spatial selection, and also the control of the specific absorption rate. © 2015 The Authors. NMR in Biomedicine published by John Wiley & Sons Ltd.© 2015 The Authors. NMR in Biomedicine published by John Wiley & Sons Ltd.
Safety and imaging performance of two-channel RF shimming for fetal MRI at 3 T
[J].
DOI:10.1002/mrm.28895
URL
[本文引用: 1]

This study investigates whether two‐channel radiofrequency (RF) shimming can improve imaging without increasing specific absorption rate (SAR) for fetal MRI at 3T.
Impacts of RF shimming on local SAR caused by MRI 3 T birdcage coil near femoral plate implants
[C]//
Statistical evaluation of radiofrequency exposure during magnetic resonant imaging: Application of whole-body individual human model and body motion in the coil
[J].
DOI:10.3390/ijerph16061069
URL
[本文引用: 1]

The accurate estimation of patient’s exposure to the radiofrequency (RF) electromagnetic field of magnetic resonance imaging (MRI) significantly depends on a precise individual anatomical model. In the study, we investigated the applicability of an efficient whole-body individual modelling method for the assessment of MRI RF exposure. The individual modelling method included a deformable human model and tissue simplification techniques. Besides its remarkable efficiency, this approach utilized only a low specific absorption rate (SAR) sequence or even no MRI scan to generate the whole-body individual model. Therefore, it substantially reduced the risk of RF exposure. The dosimetric difference of the individual modelling method was evaluated using the manually segmented human models. In addition, stochastic dosimetry using a surrogate model by polynomial chaos presented SAR variability due to body misalignment and tilt in the coil, which were frequently occurred in the practical scan. In conclusion, the dosimetric equivalence of the individual models was validated by both deterministic and stochastic dosimetry. The proposed individual modelling method allowed the physicians to quantify the patient-specific SAR while the statistical results enabled them to comprehensively weigh over the exposure risk and get the benefit of imaging enhancement by using the high-intensity scanners or the high-SAR sequences.
SAR and temperature distributions in a database of realistic human models for 7 T cardiac imaging
[J].
DOI:10.1002/nbm.4525
PMID:33955061
[本文引用: 1]

To investigate inter-subject variability of B, SAR and temperature rise in a database of human models using a local transmit array for 7 T cardiac imaging.Dixon images were acquired of 14 subjects and segmented in dielectric models with an eight-channel local transmit array positioned around the torso for cardiac imaging. EM simulations were done to calculate SAR distributions. Based on the SAR distributions, temperature simulations were performed for exposure times of 6 min and 30 min. Peak local SAR and temperature rise levels were calculated for different RF shim settings. A statistical analysis of the resulting peak local SAR and temperature rise levels was performed to arrive at safe power limits.For RF shim vectors with random phase and uniformly distributed power, a safe average power limit of 35.7 W was determined (first level controlled mode). When RF amplitude and phase shimming was performed on the heart, a safe average power limit of 35.0 W was found. According to Pennes' model, our numerical study suggests a very low probability of exceeding the absolute local temperature limit of 40 °C for a total exposure time of 6 min and a peak local SAR of 20 W/kg. For a 30 min exposure time at 20 W/kg, it was shown that the absolute temperature limit can be exceeded in the case where perfusion does not change with temperature.Safe power constraints were found for 7 T cardiac imaging with an eight-channel local transmit array, while considering the inter-subject variability of B, SAR and temperature rise.© 2021 The Authors. NMR in Biomedicine published by John Wiley & Sons Ltd.
Temperature-based MRI safety simulations with a limited number of tissues
[J].
DOI:10.1002/mrm.28693
PMID:33547673
[本文引用: 1]

Demonstrate ability to produce reasonable simulations of temperature using numerical models of the human body with a limited number of tissues.For both a male and female human body model, numerical simulations were used to calculate temperature distributions in three different models of the same human body: the original model with 35 tissues for the male model and 76 tissues for the female model, a simplified model having only three tissues (muscle, fat, and lung), and a simplified model having six tissues (muscle, fat, lung, bone, brain, and skin).Although a three-tissue model gave reasonable specific absorption rate estimates in comparison to an original with many more tissues, because of tissue-specific thermal and physiological properties that do not affect specific absorption rate, such as rate of perfusion by blood, the three-tissue model did not provide temperature distributions similar to those of the original model. Inclusion of a few additional tissues, as in the six-tissue model, produced results in much better agreement with those from the original model.Reasonable estimates of temperature can be simulated with a limited number of tissues, although this number is higher than the number of tissues required to produce reasonable simulations of specific absorption rate. For exposures primarily in the head and thorax, six tissues may be adequate for reasonable estimates of temperature.© 2021 International Society for Magnetic Resonance in Medicine.
7 T body MRI: B-1 shimming with simultaneous SAR reduction
[J].DOI:10.1088/0031-9155/52/17/022 URL [本文引用: 1]
An investigation into the minimum number of tissue groups required for 7 T in-silico parallel transmit electromagnetic safety simulations in the human head
[J].DOI:10.1002/mrm.v85.2 URL [本文引用: 1]
SAR simulations for high-field MRI: how much detail, effort, and accuracy is needed?
[J]
DOI:10.1002/mrm.24329
PMID:22611018
[本文引用: 1]

Accurate prediction of specific absorption rate (SAR) for high field MRI is necessary to best exploit its potential and guarantee safe operation. To reduce the effort (time, complexity) of SAR simulations while maintaining robust results, the minimum requirements for the creation (segmentation, labeling) of human models and methods to reduce the time for SAR calculations for 7 Tesla MR-imaging are evaluated. The geometric extent of the model required for realistic head-simulations and the number of tissue types sufficient to form a reliable but simplified model of the human body are studied. Two models (male and female) of the virtual family are analyzed. Additionally, their position within the head-coil is taken into account. Furthermore, the effects of retuning the coils to different load conditions and the influence of a large bore radiofrequency-shield have been examined. The calculation time for SAR simulations in the head can be reduced by 50% without significant error for smaller model extent and simplified tissue structure outside the coil. Likewise, the model generation can be accelerated by reducing the number of tissue types. Local SAR can vary up to 14% due to position alone. This must be considered and sets a limit for SAR prediction accuracy. All these results are comparable between the two body models tested.Copyright © 2012 Wiley Periodicals, Inc.
Multi-planar 3D knee MRI segmentation via UNet inspired architectures
[J].DOI:10.1002/ima.v33.3 URL [本文引用: 1]
Generative adversarial networks
[J].
DOI:10.1145/3422622
URL
[本文引用: 1]

\n Generative adversarial networks are a kind of artificial intelligence algorithm designed to solve the\n generative modeling\n problem. The goal of a generative model is to study a collection of training examples and learn the probability distribution that generated them. Generative Adversarial Networks (GANs) are then able to generate more examples from the estimated probability distribution. Generative models based on deep learning are common, but GANs are among the most successful generative models (especially in terms of their ability to generate realistic high-resolution images). GANs have been successfully applied to a wide variety of tasks (mostly in research settings) but continue to present unique challenges and research opportunities because they are based on game theory while most other approaches to generative modeling are based on optimization.\n
Applications of generative adversarial networks in medical image processing
[J].
生成对抗网络在医学图像转换领域的应用
[J].
Efficient and accurate MRI super-resolution using a generative adversarial network and 3D multi-level densely connected network
[C]//
Cycle-consistent adversarial denoising network for multiphase coronary CT angiography
[J].
DOI:10.1002/mp.13284
PMID:30449055
[本文引用: 1]

In multiphase coronary CT angiography (CTA), a series of CT images are taken at different levels of radiation dose during the examination. Although this reduces the total radiation dose, the image quality during the low-dose phases is significantly degraded. Recently, deep neural network approaches based on supervised learning technique have demonstrated impressive performance improvement over conventional model-based iterative methods for low-dose CT. However, matched low- and routine-dose CT image pairs are difficult to obtain in multiphase CT. To address this problem, we aim at developing a new deep learning framework.We propose an unsupervised learning technique that can remove the noise of the CT images in the low-dose phases by learning from the CT images in the routine dose phases. Although a supervised learning approach is not applicable due to the differences in the underlying heart structure in two phases, the images are closely related in two phases, so we propose a cycle-consistent adversarial denoising network to learn the mapping between the low- and high-dose cardiac phases.Experimental results showed that the proposed method effectively reduces the noise in the low-dose CT image while preserving detailed texture and edge information. Moreover, thanks to the cyclic consistency and identity loss, the proposed network does not create any artificial features that are not present in the input images. Visual grading and quality evaluation also confirm that the proposed method provides significant improvement in diagnostic quality.The proposed network can learn the image distributions from the routine-dose cardiac phases, which is a big advantage over the existing supervised learning networks that need exactly matched low- and routine-dose CT images. Considering the effectiveness and practicability of the proposed method, we believe that the proposed can be applied for many other CT acquisition protocols.© 2018 American Association of Physicists in Medicine.
Generative adversarial learning for reducing manual annotation in semantic segmentation on large scale miscroscopy images: Automated vessel segmentation in retinal fundus image as test case
[C]//
Automated cartilage and meniscus segmentation of knee MRI with conditional generative adversarial networks
[J].
DOI:10.1002/mrm.28111
PMID:31793071
[本文引用: 1]

Fully automatic tissue segmentation is an essential step to translate quantitative MRI techniques to clinical setting. The goal of this study was to develop a novel approach based on the generative adversarial networks for fully automatic segmentation of knee cartilage and meniscus.Defining proper loss function for semantic segmentation to enforce the learning of multiscale spatial constraints in an end-to-end training process is an open problem. In this work, we have used the conditional generative adversarial networks to improve segmentation performance of convolutional neural network, such as UNet alone by overcoming the problems caused by pixel-wise mapping based objective functions, and to capture cartilage features during the training of the network. Furthermore, the Dice coefficient and cross entropy losses were incorporated to the loss functions to improve the model performance. The model was trained and tested on 176, 3D DESS (double-echo steady-state) knee images from the Osteoarthritis Initiative data set.The proposed model provided excellent segmentation performance for cartilages with Dice coefficients ranging from 0.84 in patellar cartilage to 0.91 in lateral tibial cartilage, with an average Dice coefficient of 0.88. For meniscus segmentation, the model achieves 0.89 Dice coefficient for lateral meniscus and 0.87 Dice coefficient for medial meniscus. The results are superior to previously published automatic cartilage and meniscus segmentation methods based on deep learning models such as convolutional neural network.The proposed UNet-conditional generative adversarial networks based model demonstrated a fully automated segmentation method with high accuracy for knee cartilage and meniscus.© 2019 International Society for Magnetic Resonance in Medicine.
Semi-supervised assessment of incomplete LV coverage in cardiac MRI using generative adversarial nets
[C]//
Conditional generative adversarial nets
[J].
Knee joint image segmentation and model construction based on cascaded network
[J].
基于级联网络的膝关节图像分割与模型构建
[J].
Attention gated networks: Learning to leverage salient regions in medical images
[J].
Local SAR in parallel transmission pulse design
[J].
DOI:10.1002/mrm.23140
PMID:22083594
[本文引用: 2]

The management of local and global power deposition in human subjects (specific absorption rate, SAR) is a fundamental constraint to the application of parallel transmission (pTx) systems. Even though the pTx and single channel have to meet the same SAR requirements, the complex behavior of the spatial distribution of local SAR for transmission arrays poses problems that are not encountered in conventional single-channel systems and places additional requirements on pTx radio frequency pulse design. We propose a pTx pulse design method which builds on recent work to capture the spatial distribution of local SAR in numerical tissue models in a compressed parameterization in order to incorporate local SAR constraints within computation times that accommodate pTx pulse design during an in vivo magnetic resonance imaging scan. Additionally, the algorithm yields a protocol-specific ultimate peak in local SAR, which is shown to bound the achievable peak local SAR for a given excitation profile fidelity. The performance of the approach was demonstrated using a numerical human head model and a 7 Tesla eight-channel transmit array. The method reduced peak local 10 g SAR by 14-66% for slice-selective pTx excitations and 2D selective pTx excitations compared to a pTx pulse design constrained only by global SAR. The primary tradeoff incurred for reducing peak local SAR was an increase in global SAR, up to 34% for the evaluated examples, which is favorable in cases where local SAR constraints dominate the pulse applications.Copyright © 2011 Wiley Periodicals, Inc.
Individualized SAR calculations using computer vision-based MR segmentation and a fast electromagnetic solver
[J].DOI:10.1002/mrm.v85.1 URL [本文引用: 2]
Bench to bore ramifications of inter-subject head differences on RF shimming and specific absorption rates at 7 T
[J].DOI:10.1016/j.mri.2022.07.009 URL [本文引用: 1]
The Virtual Family—development of surface-based anatomical models of two adults and two children for dosimetric simulations
[J].DOI:10.1088/0031-9155/55/2/N01 URL [本文引用: 1]