NIA-AA research framework: Toward a biological definition of Alzheimer’s disease
1
2018
... 阿尔兹海默症(Alzheimer disease,AD)是痴呆症最常见的类型,占所有痴呆症的50~70%,其症状为认知、功能和行为的退化,通常始于对于最近发生事件记忆的丧失.AD患者的脑组织病理特征是脑脊液中β 淀粉蛋白(Aβ 42)水平下降,总Tau蛋白或磷酸化Tau蛋白升高,从而在细胞内聚集成神经原纤维缠结(neurofibrillary tangles,NFT)[1 ] .全球有约5 000万人患有AD,由于人口老龄化,预计到2050年,患者数量将增加两倍,即85人中就有一人患有AD,这样的趋势无疑对残疾风险、疾病负担和医疗费用是个巨大的挑战[2 ] . ...
Alzheimer’s disease
1
2021
... 阿尔兹海默症(Alzheimer disease,AD)是痴呆症最常见的类型,占所有痴呆症的50~70%,其症状为认知、功能和行为的退化,通常始于对于最近发生事件记忆的丧失.AD患者的脑组织病理特征是脑脊液中β 淀粉蛋白(Aβ 42)水平下降,总Tau蛋白或磷酸化Tau蛋白升高,从而在细胞内聚集成神经原纤维缠结(neurofibrillary tangles,NFT)[1 ] .全球有约5 000万人患有AD,由于人口老龄化,预计到2050年,患者数量将增加两倍,即85人中就有一人患有AD,这样的趋势无疑对残疾风险、疾病负担和医疗费用是个巨大的挑战[2 ] . ...
State of the science on mild cognitive impairment
1
2020
... AD的加重也不是突然的,而是存在一个很长的被称为轻度认知障碍(mild cognitive impairment,MCI)的发展阶段,MCI的早期诊断对于AD的治疗至关重要[3 ] .临床评估仍然是目前最主要的AD诊断手段,特别是针对患者本人的临床访谈,同时对患者进行神经衰退记录,辅助方法包括检测海马体积完整性、追踪白质纤维[4 ] 等.这样的评估方法非常耗时耗力,针对一个患者就需要耗费大量的医疗资源对其AD状态进行评估,如果可以开发一种省时且无创的辅助诊断方法,则可大大降低医疗资源的消耗,减少患者被漏诊的可能.随着机器学习方法被引入到医学影像分类中,许多研究者通过大量医学影像数据与机器学习算法提出了针对AD与MCI的预测方法[5 ⇓ -7 ] ,且准确率已经接近人类专家的诊断效果,但是传统的机器学习算法依赖于人工提取的特征,需要复杂的数据预处理操作.而在实际应用场景中,人们更倾向于进行较少的数据预处理步骤就可以得到可靠的结果,这样端到端的学习模式正符合深度学习的特性. ...
DTI measurements for Alzheimer’s classification
1
2017
... AD的加重也不是突然的,而是存在一个很长的被称为轻度认知障碍(mild cognitive impairment,MCI)的发展阶段,MCI的早期诊断对于AD的治疗至关重要[3 ] .临床评估仍然是目前最主要的AD诊断手段,特别是针对患者本人的临床访谈,同时对患者进行神经衰退记录,辅助方法包括检测海马体积完整性、追踪白质纤维[4 ] 等.这样的评估方法非常耗时耗力,针对一个患者就需要耗费大量的医疗资源对其AD状态进行评估,如果可以开发一种省时且无创的辅助诊断方法,则可大大降低医疗资源的消耗,减少患者被漏诊的可能.随着机器学习方法被引入到医学影像分类中,许多研究者通过大量医学影像数据与机器学习算法提出了针对AD与MCI的预测方法[5 ⇓ -7 ] ,且准确率已经接近人类专家的诊断效果,但是传统的机器学习算法依赖于人工提取的特征,需要复杂的数据预处理操作.而在实际应用场景中,人们更倾向于进行较少的数据预处理步骤就可以得到可靠的结果,这样端到端的学习模式正符合深度学习的特性. ...
Performance of machine learning methods applied to structural MRI and ADAS cognitive scores in diagnosing Alzheimer’s disease
2
2019
... AD的加重也不是突然的,而是存在一个很长的被称为轻度认知障碍(mild cognitive impairment,MCI)的发展阶段,MCI的早期诊断对于AD的治疗至关重要[3 ] .临床评估仍然是目前最主要的AD诊断手段,特别是针对患者本人的临床访谈,同时对患者进行神经衰退记录,辅助方法包括检测海马体积完整性、追踪白质纤维[4 ] 等.这样的评估方法非常耗时耗力,针对一个患者就需要耗费大量的医疗资源对其AD状态进行评估,如果可以开发一种省时且无创的辅助诊断方法,则可大大降低医疗资源的消耗,减少患者被漏诊的可能.随着机器学习方法被引入到医学影像分类中,许多研究者通过大量医学影像数据与机器学习算法提出了针对AD与MCI的预测方法[5 ⇓ -7 ] ,且准确率已经接近人类专家的诊断效果,但是传统的机器学习算法依赖于人工提取的特征,需要复杂的数据预处理操作.而在实际应用场景中,人们更倾向于进行较少的数据预处理步骤就可以得到可靠的结果,这样端到端的学习模式正符合深度学习的特性. ...
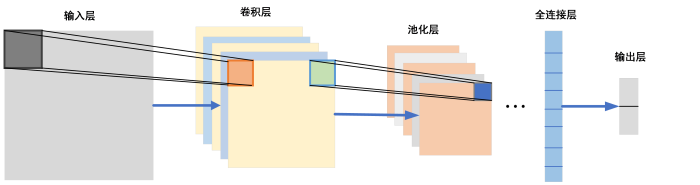
... 以往用于AD影像分类的传统机器学习算法,如支持向量机[12 ] 、决策树[13 ] 和随机森林[14 ] 等,均依赖于人工对图像特征进行提取:通常需要先使用脑模板,例如自动解剖标记(automated anatomical labeling,AAL),手动划分感兴趣区域(region of interest,ROI),然后从ROI中提取灰度直方图[14 ] 、灰质体积、皮层表面积[12 ] 、皮层分形结构[5 ] 、海马体体积[7 ] 等作为图像特征.该类特征提取方法依赖于专家的先验知识,同时手动划分的ROI具有不可避免的人为误差,从而影响到机器学习算法的表现.而深度学习端到端学习的特性解决了这样的难题,只需进行简单的预处理,例如将图像进行配准、归一化和平滑后直接作为矩阵输入到模型中,就可以自动进行特征提取,然后使用全连接层或者传统机器学习方法作为分类器.在实际的临床应用场景中,不同的医院以及不同的采集设备都会影响到成像效果,而不依赖于人工图像特征提取的深度学习算法往往会得到更好的效果,而且泛化能力也可以得到保证. ...
Volumetric histogram-based Alzheimer’s disease detection using support vector machine
1
2019
... AD的加重也不是突然的,而是存在一个很长的被称为轻度认知障碍(mild cognitive impairment,MCI)的发展阶段,MCI的早期诊断对于AD的治疗至关重要[3 ] .临床评估仍然是目前最主要的AD诊断手段,特别是针对患者本人的临床访谈,同时对患者进行神经衰退记录,辅助方法包括检测海马体积完整性、追踪白质纤维[4 ] 等.这样的评估方法非常耗时耗力,针对一个患者就需要耗费大量的医疗资源对其AD状态进行评估,如果可以开发一种省时且无创的辅助诊断方法,则可大大降低医疗资源的消耗,减少患者被漏诊的可能.随着机器学习方法被引入到医学影像分类中,许多研究者通过大量医学影像数据与机器学习算法提出了针对AD与MCI的预测方法[5 ⇓ -7 ] ,且准确率已经接近人类专家的诊断效果,但是传统的机器学习算法依赖于人工提取的特征,需要复杂的数据预处理操作.而在实际应用场景中,人们更倾向于进行较少的数据预处理步骤就可以得到可靠的结果,这样端到端的学习模式正符合深度学习的特性. ...
Hippocampal atrophy based Alzheimer’s disease diagnosis via machine learning methods
2
2020
... AD的加重也不是突然的,而是存在一个很长的被称为轻度认知障碍(mild cognitive impairment,MCI)的发展阶段,MCI的早期诊断对于AD的治疗至关重要[3 ] .临床评估仍然是目前最主要的AD诊断手段,特别是针对患者本人的临床访谈,同时对患者进行神经衰退记录,辅助方法包括检测海马体积完整性、追踪白质纤维[4 ] 等.这样的评估方法非常耗时耗力,针对一个患者就需要耗费大量的医疗资源对其AD状态进行评估,如果可以开发一种省时且无创的辅助诊断方法,则可大大降低医疗资源的消耗,减少患者被漏诊的可能.随着机器学习方法被引入到医学影像分类中,许多研究者通过大量医学影像数据与机器学习算法提出了针对AD与MCI的预测方法[5 ⇓ -7 ] ,且准确率已经接近人类专家的诊断效果,但是传统的机器学习算法依赖于人工提取的特征,需要复杂的数据预处理操作.而在实际应用场景中,人们更倾向于进行较少的数据预处理步骤就可以得到可靠的结果,这样端到端的学习模式正符合深度学习的特性. ...
... 以往用于AD影像分类的传统机器学习算法,如支持向量机[12 ] 、决策树[13 ] 和随机森林[14 ] 等,均依赖于人工对图像特征进行提取:通常需要先使用脑模板,例如自动解剖标记(automated anatomical labeling,AAL),手动划分感兴趣区域(region of interest,ROI),然后从ROI中提取灰度直方图[14 ] 、灰质体积、皮层表面积[12 ] 、皮层分形结构[5 ] 、海马体体积[7 ] 等作为图像特征.该类特征提取方法依赖于专家的先验知识,同时手动划分的ROI具有不可避免的人为误差,从而影响到机器学习算法的表现.而深度学习端到端学习的特性解决了这样的难题,只需进行简单的预处理,例如将图像进行配准、归一化和平滑后直接作为矩阵输入到模型中,就可以自动进行特征提取,然后使用全连接层或者传统机器学习方法作为分类器.在实际的临床应用场景中,不同的医院以及不同的采集设备都会影响到成像效果,而不依赖于人工图像特征提取的深度学习算法往往会得到更好的效果,而且泛化能力也可以得到保证. ...
基于计算机辅助诊断技术的阿尔兹海默症早期分类研究综述
1
2022
... 在基于机器学习的AD影像学分类领域,以往的综述论文往往从机器学习和深度学习两大方面进行讨论[8 ,9 ] ,或者着重于特定算法在AD影像学分类中的应用[10 ,11 ] .而本文针对深度学习的讨论超出特定算法层面,着重于迁移学习、集成学习和多任务学习这些方法对基于深度学习的AD影像学分类效能的提升;针对深度学习的黑箱特性所带来的可解释性难题,本文对目前可解释性的研究进展也做了分类探讨;同时还讨论了脑龄预测任务与AD影像学分类任务的相关性,这一方向未来或许会成为该领域新的研究重点. ...
基于计算机辅助诊断技术的阿尔兹海默症早期分类研究综述
1
2022
... 在基于机器学习的AD影像学分类领域,以往的综述论文往往从机器学习和深度学习两大方面进行讨论[8 ,9 ] ,或者着重于特定算法在AD影像学分类中的应用[10 ,11 ] .而本文针对深度学习的讨论超出特定算法层面,着重于迁移学习、集成学习和多任务学习这些方法对基于深度学习的AD影像学分类效能的提升;针对深度学习的黑箱特性所带来的可解释性难题,本文对目前可解释性的研究进展也做了分类探讨;同时还讨论了脑龄预测任务与AD影像学分类任务的相关性,这一方向未来或许会成为该领域新的研究重点. ...
基于智能影像基因组学技术的阿尔茨海默病预测进展
1
2021
... 在基于机器学习的AD影像学分类领域,以往的综述论文往往从机器学习和深度学习两大方面进行讨论[8 ,9 ] ,或者着重于特定算法在AD影像学分类中的应用[10 ,11 ] .而本文针对深度学习的讨论超出特定算法层面,着重于迁移学习、集成学习和多任务学习这些方法对基于深度学习的AD影像学分类效能的提升;针对深度学习的黑箱特性所带来的可解释性难题,本文对目前可解释性的研究进展也做了分类探讨;同时还讨论了脑龄预测任务与AD影像学分类任务的相关性,这一方向未来或许会成为该领域新的研究重点. ...
基于智能影像基因组学技术的阿尔茨海默病预测进展
1
2021
... 在基于机器学习的AD影像学分类领域,以往的综述论文往往从机器学习和深度学习两大方面进行讨论[8 ,9 ] ,或者着重于特定算法在AD影像学分类中的应用[10 ,11 ] .而本文针对深度学习的讨论超出特定算法层面,着重于迁移学习、集成学习和多任务学习这些方法对基于深度学习的AD影像学分类效能的提升;针对深度学习的黑箱特性所带来的可解释性难题,本文对目前可解释性的研究进展也做了分类探讨;同时还讨论了脑龄预测任务与AD影像学分类任务的相关性,这一方向未来或许会成为该领域新的研究重点. ...
深度学习方法在阿尔茨海默病脑图像方面的应用进展
1
2019
... 在基于机器学习的AD影像学分类领域,以往的综述论文往往从机器学习和深度学习两大方面进行讨论[8 ,9 ] ,或者着重于特定算法在AD影像学分类中的应用[10 ,11 ] .而本文针对深度学习的讨论超出特定算法层面,着重于迁移学习、集成学习和多任务学习这些方法对基于深度学习的AD影像学分类效能的提升;针对深度学习的黑箱特性所带来的可解释性难题,本文对目前可解释性的研究进展也做了分类探讨;同时还讨论了脑龄预测任务与AD影像学分类任务的相关性,这一方向未来或许会成为该领域新的研究重点. ...
深度学习方法在阿尔茨海默病脑图像方面的应用进展
1
2019
... 在基于机器学习的AD影像学分类领域,以往的综述论文往往从机器学习和深度学习两大方面进行讨论[8 ,9 ] ,或者着重于特定算法在AD影像学分类中的应用[10 ,11 ] .而本文针对深度学习的讨论超出特定算法层面,着重于迁移学习、集成学习和多任务学习这些方法对基于深度学习的AD影像学分类效能的提升;针对深度学习的黑箱特性所带来的可解释性难题,本文对目前可解释性的研究进展也做了分类探讨;同时还讨论了脑龄预测任务与AD影像学分类任务的相关性,这一方向未来或许会成为该领域新的研究重点. ...
Machine learning techniques for the diagnosis of Alzheimer’s disease: a review
1
2020
... 在基于机器学习的AD影像学分类领域,以往的综述论文往往从机器学习和深度学习两大方面进行讨论[8 ,9 ] ,或者着重于特定算法在AD影像学分类中的应用[10 ,11 ] .而本文针对深度学习的讨论超出特定算法层面,着重于迁移学习、集成学习和多任务学习这些方法对基于深度学习的AD影像学分类效能的提升;针对深度学习的黑箱特性所带来的可解释性难题,本文对目前可解释性的研究进展也做了分类探讨;同时还讨论了脑龄预测任务与AD影像学分类任务的相关性,这一方向未来或许会成为该领域新的研究重点. ...
Classification of Alzheimer’s disease based on brain MRI and machine learning
2
2020
... 以往用于AD影像分类的传统机器学习算法,如支持向量机[12 ] 、决策树[13 ] 和随机森林[14 ] 等,均依赖于人工对图像特征进行提取:通常需要先使用脑模板,例如自动解剖标记(automated anatomical labeling,AAL),手动划分感兴趣区域(region of interest,ROI),然后从ROI中提取灰度直方图[14 ] 、灰质体积、皮层表面积[12 ] 、皮层分形结构[5 ] 、海马体体积[7 ] 等作为图像特征.该类特征提取方法依赖于专家的先验知识,同时手动划分的ROI具有不可避免的人为误差,从而影响到机器学习算法的表现.而深度学习端到端学习的特性解决了这样的难题,只需进行简单的预处理,例如将图像进行配准、归一化和平滑后直接作为矩阵输入到模型中,就可以自动进行特征提取,然后使用全连接层或者传统机器学习方法作为分类器.在实际的临床应用场景中,不同的医院以及不同的采集设备都会影响到成像效果,而不依赖于人工图像特征提取的深度学习算法往往会得到更好的效果,而且泛化能力也可以得到保证. ...
... [12 ]、皮层分形结构[5 ] 、海马体体积[7 ] 等作为图像特征.该类特征提取方法依赖于专家的先验知识,同时手动划分的ROI具有不可避免的人为误差,从而影响到机器学习算法的表现.而深度学习端到端学习的特性解决了这样的难题,只需进行简单的预处理,例如将图像进行配准、归一化和平滑后直接作为矩阵输入到模型中,就可以自动进行特征提取,然后使用全连接层或者传统机器学习方法作为分类器.在实际的临床应用场景中,不同的医院以及不同的采集设备都会影响到成像效果,而不依赖于人工图像特征提取的深度学习算法往往会得到更好的效果,而且泛化能力也可以得到保证. ...
An optimized decision tree with genetic algorithm rule-based approach to reveal the brain’s changes during Alzheimer’s disease dementia
1
2021
... 以往用于AD影像分类的传统机器学习算法,如支持向量机[12 ] 、决策树[13 ] 和随机森林[14 ] 等,均依赖于人工对图像特征进行提取:通常需要先使用脑模板,例如自动解剖标记(automated anatomical labeling,AAL),手动划分感兴趣区域(region of interest,ROI),然后从ROI中提取灰度直方图[14 ] 、灰质体积、皮层表面积[12 ] 、皮层分形结构[5 ] 、海马体体积[7 ] 等作为图像特征.该类特征提取方法依赖于专家的先验知识,同时手动划分的ROI具有不可避免的人为误差,从而影响到机器学习算法的表现.而深度学习端到端学习的特性解决了这样的难题,只需进行简单的预处理,例如将图像进行配准、归一化和平滑后直接作为矩阵输入到模型中,就可以自动进行特征提取,然后使用全连接层或者传统机器学习方法作为分类器.在实际的临床应用场景中,不同的医院以及不同的采集设备都会影响到成像效果,而不依赖于人工图像特征提取的深度学习算法往往会得到更好的效果,而且泛化能力也可以得到保证. ...
Automatic detection of Alzheimer disease based on histogram and random forest
2
2019
... 以往用于AD影像分类的传统机器学习算法,如支持向量机[12 ] 、决策树[13 ] 和随机森林[14 ] 等,均依赖于人工对图像特征进行提取:通常需要先使用脑模板,例如自动解剖标记(automated anatomical labeling,AAL),手动划分感兴趣区域(region of interest,ROI),然后从ROI中提取灰度直方图[14 ] 、灰质体积、皮层表面积[12 ] 、皮层分形结构[5 ] 、海马体体积[7 ] 等作为图像特征.该类特征提取方法依赖于专家的先验知识,同时手动划分的ROI具有不可避免的人为误差,从而影响到机器学习算法的表现.而深度学习端到端学习的特性解决了这样的难题,只需进行简单的预处理,例如将图像进行配准、归一化和平滑后直接作为矩阵输入到模型中,就可以自动进行特征提取,然后使用全连接层或者传统机器学习方法作为分类器.在实际的临床应用场景中,不同的医院以及不同的采集设备都会影响到成像效果,而不依赖于人工图像特征提取的深度学习算法往往会得到更好的效果,而且泛化能力也可以得到保证. ...
... [14 ]、灰质体积、皮层表面积[12 ] 、皮层分形结构[5 ] 、海马体体积[7 ] 等作为图像特征.该类特征提取方法依赖于专家的先验知识,同时手动划分的ROI具有不可避免的人为误差,从而影响到机器学习算法的表现.而深度学习端到端学习的特性解决了这样的难题,只需进行简单的预处理,例如将图像进行配准、归一化和平滑后直接作为矩阵输入到模型中,就可以自动进行特征提取,然后使用全连接层或者传统机器学习方法作为分类器.在实际的临床应用场景中,不同的医院以及不同的采集设备都会影响到成像效果,而不依赖于人工图像特征提取的深度学习算法往往会得到更好的效果,而且泛化能力也可以得到保证. ...
Robust hybrid deep learning models for Alzheimer’s progression detection
3
2021
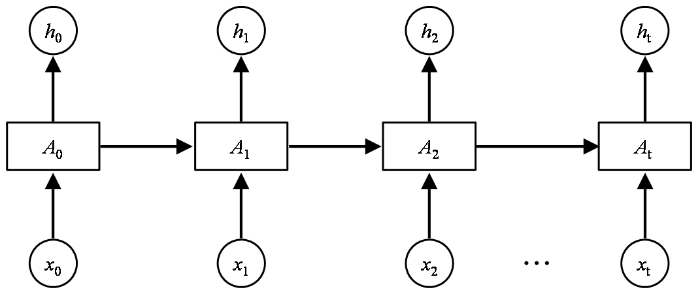
... RNN具有CNN不具备的记忆性,适用于对序列非线性特征的学习,所以通常被应用于自然语言处理领域.虽然RNN不常被应用于计算机视觉领域,但由于AD是长期发展的疾病,具有明显的时间序列性,所以依然有少量研究者将RNN应用于AD影像学分类任务中.如Abuhmed等[15 ] 使用RNN学习受试者在过去时间上的影像特征,对该受试者未来的状况进行预测.总体而言,RNN在AD影像学分类中应用不如CNN广泛. ...
... 在对AD患者进行诊断时,认知评分量表的分数是重要的参考标准[44 ] ,也有很多AD影像分类的论文[45 ] 将MMSE得分作为AD、MCI和HC组的标签,所以对于认知评分的回归预测也是AD影像分类多任务学习的理想辅助任务.如Zeng等[46 ] 将MMSE和AD评定量表-认知分量表(Alzheimer’s disease assessment scale,ADAS-cog)的回归预测作为辅助任务,使用深度信念网络(deep belief network,DBN)作为网络主干,最终得到的网络在AD与HC的分类、pMCI与HC的分类和AD与sMCI的分类中的准确率均超过95%.Abuhmed等[15 ] 使用患者在基线,以及基线后6个月、12个月和18个月的PET、sMRI、神经心理学、神经病理学和认知评分多模态数据,利用BiLSTM对患者基线后48个月的CDR和MMSE等7个认知评分进行回归预测,将预测得到的7个认知评分和患者的年龄、性别等信息作为随机森林分类器的输入,最后对患者基线48个月之后的状态进行AD、MCI与HC三分类,准确率达到了84.95%. ...
... Performance comparison of different models
Table 2 第一作者 分类任务 数据模态 数据集 测试方法 分类模型 准确率% 郁松[21 ] AD/HC sMRI 1015 HC 训练集60% 3D ResNet-101 97.425 Parmar[24 ] AD/HC fMRI 30 AD 训练集60% 3D CNN 94.58 Bi[19 ] AD/HC fMRI 118 AD 五折交叉验证 RNN,ELM 91.3 (AUC) Yi̇Ği̇T[20 ] AD/HC sMRI 训练集 测试集 / 2D CNN 83 Punjabi[33 ] AD/HC PET 共1299 / 3D CNN 85.15 Zhang[49 ] AD/HC sMRI 200 AD 五折交叉验证 3D ResAttNet 91.3 Qiu[52 ] AD/HC sMRI+性别+年龄+MMSE / 训练集60% FCN 96.8 Deng[28 ] AD/HC DTI+sMRI 98 AD 训练集60% CNN 90.00 Marzban[29 ] AD/HC DTI+sMRI 115 AD 十折交叉验证 CNN 93.5 Kang[30 ] EMCI/HC DTI+sMRI 50 HC 训练集80% VGG-16,LASSO 94.2 Kwak[54 ] AD/HC sMRI 110 AD 五折交叉验证 DenseNet 93.75 Ju[16 ] MCI/HC fMRI 91 MCI 十折交叉验证 AE 86.47 Baydargil[17 ] AD/MCI/HC PET 141 AD 训练集80% CAE 98.67 Guan[51 ] AD/HC sMRI 384 AD 训练集 90% ResNet、pABN 90.7 Li[34 ] AD/MCI/HC MRI 237 AD 训练集65% ResNet-200 83 Basaia[40 ] AD/HC sMRI 294 AD 训练集90% 3D FCN(迁移学习) 99.2 孔伶旭[36 ] EMCI/HC fMRI 32 HC 五折交叉验证 Mobilenet(迁移学习) 73.67 Bin[37 ] AD/HC sMRI 100 AD 五折交叉验证 Inception-v3 99.45 Massalimova[27 ] AD/MCI/HC DTI 训练集 测试集 / ResNet-18(迁移学习) 97 Mehmood[38 ] AD/HC sMRI 75 AD 训练集64% VGG-19(迁移学习) 95.33 Raju[50 ] 非常轻度痴呆/轻度痴呆/中度痴呆/HC(四分类) sMRI 1013非常轻度痴呆 334非常轻度痴呆 VGG-16(迁移学习) 99 Lian[39 ] AD/HC sMRI 数据集1 数据集2 两个数据集间 H-FCN(迁移学习) 90.3 Oh[18 ] AD/HC sMRI 198 AD 五折交叉验证 3D CNN,ICAE 88.6 金祝新[53 ] AD/MCI sMRI 267 AD 训练集90% 3D CNN(迁移学习) 94.6 曾安[42 ] AD/HC sMRI 137 AD 五折交叉验证 2D CNN(集成学习) 81 Bi[22 ] AD/HC sMRI 243 AD / PCANet,k -means 89.15 Choi[41 ] AD/HC sMRI 715 AD 训练集60% VGG-16,GoogLeNet,AlexNet(集成学习) 93.84 Venugopalan[55 ] AD/HC sMRI+电子病历+基因数据 共220 十折交叉验证 AE,CNN,随机森林 88 Zeng[46 ] sMCI/pMCI sMRI 92 AD 训练集70% PCA,DBN(多任务) 87.78 Spasov[43 ] sMCI/pMCI sMRI 192 AD 十折交叉验证 CNN(多任务) 86 Liu[48 ] AD/HC sMRI 97 AD 五折交叉验证 V-Net,DenseNet 88.9 Abuhmed[15 ] AD/MCI/HC PET+sMRI+神经心理学数据+神经病理学数据+认知评分 共1371 十折交叉验证 BiLSTM,随机森林 84.95
10.11938/cjmr20223013.T0003 表3 可解释性方法及AD相关脑区 ...
Early diagnosis of Alzheimer’s disease based on resting-state brain networks and deep learning
2
2019
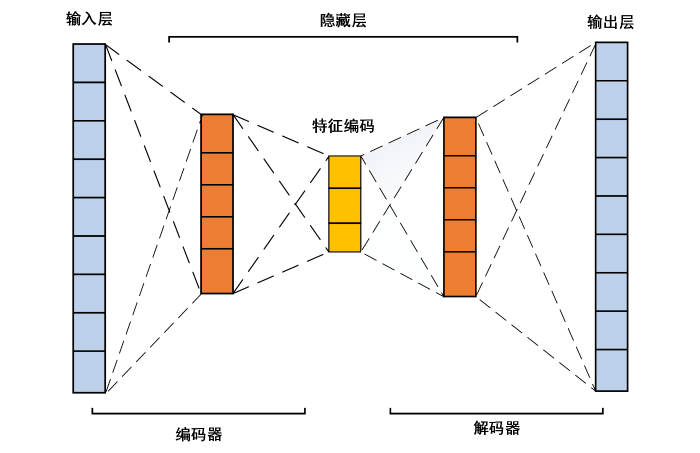
... AE作为一种无监督学习模型,不需要对训练样本打上标签,降低了获取数据的难度,所以也常被用于医学图像处理.如Ju等[16 ] 使用AE对图像特征进行提取后分类,使MCI与健康对照组(healthy control,HC)的分类准确率达到86.47%,相比传统机器学习方法提高了近20%.对于端到端的学习方式,需要使用基于AE改进的卷积自动编码机(convolutional autoencoder,CAE)对图像特征进行提取.如Baydargil等[17 ] 使用CAE进行特征提取,使AD、MCI和HC的三分类准确率达到98.67%;Oh等[18 ] 使用带有Inception多尺度卷积模块的CAE(inception modal based convolutional autoencoder,ICAE),使AD和HC的分类准确率达到88.6%. ...
... Performance comparison of different models
Table 2 第一作者 分类任务 数据模态 数据集 测试方法 分类模型 准确率% 郁松[21 ] AD/HC sMRI 1015 HC 训练集60% 3D ResNet-101 97.425 Parmar[24 ] AD/HC fMRI 30 AD 训练集60% 3D CNN 94.58 Bi[19 ] AD/HC fMRI 118 AD 五折交叉验证 RNN,ELM 91.3 (AUC) Yi̇Ği̇T[20 ] AD/HC sMRI 训练集 测试集 / 2D CNN 83 Punjabi[33 ] AD/HC PET 共1299 / 3D CNN 85.15 Zhang[49 ] AD/HC sMRI 200 AD 五折交叉验证 3D ResAttNet 91.3 Qiu[52 ] AD/HC sMRI+性别+年龄+MMSE / 训练集60% FCN 96.8 Deng[28 ] AD/HC DTI+sMRI 98 AD 训练集60% CNN 90.00 Marzban[29 ] AD/HC DTI+sMRI 115 AD 十折交叉验证 CNN 93.5 Kang[30 ] EMCI/HC DTI+sMRI 50 HC 训练集80% VGG-16,LASSO 94.2 Kwak[54 ] AD/HC sMRI 110 AD 五折交叉验证 DenseNet 93.75 Ju[16 ] MCI/HC fMRI 91 MCI 十折交叉验证 AE 86.47 Baydargil[17 ] AD/MCI/HC PET 141 AD 训练集80% CAE 98.67 Guan[51 ] AD/HC sMRI 384 AD 训练集 90% ResNet、pABN 90.7 Li[34 ] AD/MCI/HC MRI 237 AD 训练集65% ResNet-200 83 Basaia[40 ] AD/HC sMRI 294 AD 训练集90% 3D FCN(迁移学习) 99.2 孔伶旭[36 ] EMCI/HC fMRI 32 HC 五折交叉验证 Mobilenet(迁移学习) 73.67 Bin[37 ] AD/HC sMRI 100 AD 五折交叉验证 Inception-v3 99.45 Massalimova[27 ] AD/MCI/HC DTI 训练集 测试集 / ResNet-18(迁移学习) 97 Mehmood[38 ] AD/HC sMRI 75 AD 训练集64% VGG-19(迁移学习) 95.33 Raju[50 ] 非常轻度痴呆/轻度痴呆/中度痴呆/HC(四分类) sMRI 1013非常轻度痴呆 334非常轻度痴呆 VGG-16(迁移学习) 99 Lian[39 ] AD/HC sMRI 数据集1 数据集2 两个数据集间 H-FCN(迁移学习) 90.3 Oh[18 ] AD/HC sMRI 198 AD 五折交叉验证 3D CNN,ICAE 88.6 金祝新[53 ] AD/MCI sMRI 267 AD 训练集90% 3D CNN(迁移学习) 94.6 曾安[42 ] AD/HC sMRI 137 AD 五折交叉验证 2D CNN(集成学习) 81 Bi[22 ] AD/HC sMRI 243 AD / PCANet,k -means 89.15 Choi[41 ] AD/HC sMRI 715 AD 训练集60% VGG-16,GoogLeNet,AlexNet(集成学习) 93.84 Venugopalan[55 ] AD/HC sMRI+电子病历+基因数据 共220 十折交叉验证 AE,CNN,随机森林 88 Zeng[46 ] sMCI/pMCI sMRI 92 AD 训练集70% PCA,DBN(多任务) 87.78 Spasov[43 ] sMCI/pMCI sMRI 192 AD 十折交叉验证 CNN(多任务) 86 Liu[48 ] AD/HC sMRI 97 AD 五折交叉验证 V-Net,DenseNet 88.9 Abuhmed[15 ] AD/MCI/HC PET+sMRI+神经心理学数据+神经病理学数据+认知评分 共1371 十折交叉验证 BiLSTM,随机森林 84.95
10.11938/cjmr20223013.T0003 表3 可解释性方法及AD相关脑区 ...
Classification of Alzheimer’s disease using stacked sparse convolutional autoencoder
2
2019
... AE作为一种无监督学习模型,不需要对训练样本打上标签,降低了获取数据的难度,所以也常被用于医学图像处理.如Ju等[16 ] 使用AE对图像特征进行提取后分类,使MCI与健康对照组(healthy control,HC)的分类准确率达到86.47%,相比传统机器学习方法提高了近20%.对于端到端的学习方式,需要使用基于AE改进的卷积自动编码机(convolutional autoencoder,CAE)对图像特征进行提取.如Baydargil等[17 ] 使用CAE进行特征提取,使AD、MCI和HC的三分类准确率达到98.67%;Oh等[18 ] 使用带有Inception多尺度卷积模块的CAE(inception modal based convolutional autoencoder,ICAE),使AD和HC的分类准确率达到88.6%. ...
... Performance comparison of different models
Table 2 第一作者 分类任务 数据模态 数据集 测试方法 分类模型 准确率% 郁松[21 ] AD/HC sMRI 1015 HC 训练集60% 3D ResNet-101 97.425 Parmar[24 ] AD/HC fMRI 30 AD 训练集60% 3D CNN 94.58 Bi[19 ] AD/HC fMRI 118 AD 五折交叉验证 RNN,ELM 91.3 (AUC) Yi̇Ği̇T[20 ] AD/HC sMRI 训练集 测试集 / 2D CNN 83 Punjabi[33 ] AD/HC PET 共1299 / 3D CNN 85.15 Zhang[49 ] AD/HC sMRI 200 AD 五折交叉验证 3D ResAttNet 91.3 Qiu[52 ] AD/HC sMRI+性别+年龄+MMSE / 训练集60% FCN 96.8 Deng[28 ] AD/HC DTI+sMRI 98 AD 训练集60% CNN 90.00 Marzban[29 ] AD/HC DTI+sMRI 115 AD 十折交叉验证 CNN 93.5 Kang[30 ] EMCI/HC DTI+sMRI 50 HC 训练集80% VGG-16,LASSO 94.2 Kwak[54 ] AD/HC sMRI 110 AD 五折交叉验证 DenseNet 93.75 Ju[16 ] MCI/HC fMRI 91 MCI 十折交叉验证 AE 86.47 Baydargil[17 ] AD/MCI/HC PET 141 AD 训练集80% CAE 98.67 Guan[51 ] AD/HC sMRI 384 AD 训练集 90% ResNet、pABN 90.7 Li[34 ] AD/MCI/HC MRI 237 AD 训练集65% ResNet-200 83 Basaia[40 ] AD/HC sMRI 294 AD 训练集90% 3D FCN(迁移学习) 99.2 孔伶旭[36 ] EMCI/HC fMRI 32 HC 五折交叉验证 Mobilenet(迁移学习) 73.67 Bin[37 ] AD/HC sMRI 100 AD 五折交叉验证 Inception-v3 99.45 Massalimova[27 ] AD/MCI/HC DTI 训练集 测试集 / ResNet-18(迁移学习) 97 Mehmood[38 ] AD/HC sMRI 75 AD 训练集64% VGG-19(迁移学习) 95.33 Raju[50 ] 非常轻度痴呆/轻度痴呆/中度痴呆/HC(四分类) sMRI 1013非常轻度痴呆 334非常轻度痴呆 VGG-16(迁移学习) 99 Lian[39 ] AD/HC sMRI 数据集1 数据集2 两个数据集间 H-FCN(迁移学习) 90.3 Oh[18 ] AD/HC sMRI 198 AD 五折交叉验证 3D CNN,ICAE 88.6 金祝新[53 ] AD/MCI sMRI 267 AD 训练集90% 3D CNN(迁移学习) 94.6 曾安[42 ] AD/HC sMRI 137 AD 五折交叉验证 2D CNN(集成学习) 81 Bi[22 ] AD/HC sMRI 243 AD / PCANet,k -means 89.15 Choi[41 ] AD/HC sMRI 715 AD 训练集60% VGG-16,GoogLeNet,AlexNet(集成学习) 93.84 Venugopalan[55 ] AD/HC sMRI+电子病历+基因数据 共220 十折交叉验证 AE,CNN,随机森林 88 Zeng[46 ] sMCI/pMCI sMRI 92 AD 训练集70% PCA,DBN(多任务) 87.78 Spasov[43 ] sMCI/pMCI sMRI 192 AD 十折交叉验证 CNN(多任务) 86 Liu[48 ] AD/HC sMRI 97 AD 五折交叉验证 V-Net,DenseNet 88.9 Abuhmed[15 ] AD/MCI/HC PET+sMRI+神经心理学数据+神经病理学数据+认知评分 共1371 十折交叉验证 BiLSTM,随机森林 84.95
10.11938/cjmr20223013.T0003 表3 可解释性方法及AD相关脑区 ...
Classification and visualization of Alzheimer’s disease using volumetric convolutional neural network and transfer learning
4
2019
... AE作为一种无监督学习模型,不需要对训练样本打上标签,降低了获取数据的难度,所以也常被用于医学图像处理.如Ju等[16 ] 使用AE对图像特征进行提取后分类,使MCI与健康对照组(healthy control,HC)的分类准确率达到86.47%,相比传统机器学习方法提高了近20%.对于端到端的学习方式,需要使用基于AE改进的卷积自动编码机(convolutional autoencoder,CAE)对图像特征进行提取.如Baydargil等[17 ] 使用CAE进行特征提取,使AD、MCI和HC的三分类准确率达到98.67%;Oh等[18 ] 使用带有Inception多尺度卷积模块的CAE(inception modal based convolutional autoencoder,ICAE),使AD和HC的分类准确率达到88.6%. ...
... Zhang等[49 ] 使用Grad-CAM对带有注意力机制的ResNet网络(3D ResAttNet)在AD与HC分类任务上进行可视化,热力图中突出显示了海马体、侧脑室和大部分皮质区域.Raju等[50 ] 使用Grad-CAM方法对认知障碍分类网络进行可视化操作,发现海马体、杏仁核和顶叶区域得到了最大程度的激活.Oh等[18 ] 使用ICAE对AD影像进行分类,并且使用类显著映射(class saliency visualization,CSV)手段,发现pMCI与sMCI的分类任务更注重于左侧杏仁核、角回和楔前回,而AD与HC分类任务更注重内侧颞叶周围、左侧海马体.Guan等[51 ] 结合了ResNet与并行注意力增强双线性网络(parallel attention-augmented bilinear network,pABN),使用CAM发现AD与HC分类、pMCI与sMCI分类的重点区域基本相似,集中于海马体、杏仁核、脑室、额叶、颞下回、颞上沟、顶枕沟和外侧裂. ...
... Performance comparison of different models
Table 2 第一作者 分类任务 数据模态 数据集 测试方法 分类模型 准确率% 郁松[21 ] AD/HC sMRI 1015 HC 训练集60% 3D ResNet-101 97.425 Parmar[24 ] AD/HC fMRI 30 AD 训练集60% 3D CNN 94.58 Bi[19 ] AD/HC fMRI 118 AD 五折交叉验证 RNN,ELM 91.3 (AUC) Yi̇Ği̇T[20 ] AD/HC sMRI 训练集 测试集 / 2D CNN 83 Punjabi[33 ] AD/HC PET 共1299 / 3D CNN 85.15 Zhang[49 ] AD/HC sMRI 200 AD 五折交叉验证 3D ResAttNet 91.3 Qiu[52 ] AD/HC sMRI+性别+年龄+MMSE / 训练集60% FCN 96.8 Deng[28 ] AD/HC DTI+sMRI 98 AD 训练集60% CNN 90.00 Marzban[29 ] AD/HC DTI+sMRI 115 AD 十折交叉验证 CNN 93.5 Kang[30 ] EMCI/HC DTI+sMRI 50 HC 训练集80% VGG-16,LASSO 94.2 Kwak[54 ] AD/HC sMRI 110 AD 五折交叉验证 DenseNet 93.75 Ju[16 ] MCI/HC fMRI 91 MCI 十折交叉验证 AE 86.47 Baydargil[17 ] AD/MCI/HC PET 141 AD 训练集80% CAE 98.67 Guan[51 ] AD/HC sMRI 384 AD 训练集 90% ResNet、pABN 90.7 Li[34 ] AD/MCI/HC MRI 237 AD 训练集65% ResNet-200 83 Basaia[40 ] AD/HC sMRI 294 AD 训练集90% 3D FCN(迁移学习) 99.2 孔伶旭[36 ] EMCI/HC fMRI 32 HC 五折交叉验证 Mobilenet(迁移学习) 73.67 Bin[37 ] AD/HC sMRI 100 AD 五折交叉验证 Inception-v3 99.45 Massalimova[27 ] AD/MCI/HC DTI 训练集 测试集 / ResNet-18(迁移学习) 97 Mehmood[38 ] AD/HC sMRI 75 AD 训练集64% VGG-19(迁移学习) 95.33 Raju[50 ] 非常轻度痴呆/轻度痴呆/中度痴呆/HC(四分类) sMRI 1013非常轻度痴呆 334非常轻度痴呆 VGG-16(迁移学习) 99 Lian[39 ] AD/HC sMRI 数据集1 数据集2 两个数据集间 H-FCN(迁移学习) 90.3 Oh[18 ] AD/HC sMRI 198 AD 五折交叉验证 3D CNN,ICAE 88.6 金祝新[53 ] AD/MCI sMRI 267 AD 训练集90% 3D CNN(迁移学习) 94.6 曾安[42 ] AD/HC sMRI 137 AD 五折交叉验证 2D CNN(集成学习) 81 Bi[22 ] AD/HC sMRI 243 AD / PCANet,k -means 89.15 Choi[41 ] AD/HC sMRI 715 AD 训练集60% VGG-16,GoogLeNet,AlexNet(集成学习) 93.84 Venugopalan[55 ] AD/HC sMRI+电子病历+基因数据 共220 十折交叉验证 AE,CNN,随机森林 88 Zeng[46 ] sMCI/pMCI sMRI 92 AD 训练集70% PCA,DBN(多任务) 87.78 Spasov[43 ] sMCI/pMCI sMRI 192 AD 十折交叉验证 CNN(多任务) 86 Liu[48 ] AD/HC sMRI 97 AD 五折交叉验证 V-Net,DenseNet 88.9 Abuhmed[15 ] AD/MCI/HC PET+sMRI+神经心理学数据+神经病理学数据+认知评分 共1371 十折交叉验证 BiLSTM,随机森林 84.95
10.11938/cjmr20223013.T0003 表3 可解释性方法及AD相关脑区 ...
... Comparison of different interpretation methods and AD-related brain regions
Table 3 第一作者 任务 分类模型 准确率% 解释性方法 脑区 Guan[51 ] AD/HC ResNet、pABN 90.7 CAM 海马体、杏仁核、脑室、额叶、颞下回、颞上沟、顶枕沟、外侧裂 Zhang[49 ] AD/HC 3D-ResAttNet 91.3 Grad-CAM 海马体、侧脑室、大部分皮质 Raju[50 ] 轻度痴呆/非常轻度痴呆/中度痴呆/HC(四分类) VGG-16(迁移学习) 99 Grad-CAM 海马体、杏仁核、顶叶 Oh[18 ] AD/HC 3D-CNN,ICAE 86.6 CSV 内侧颞叶周围、左侧海马体 Qiu[52 ] AD/HC FCN 96.8 基于斑块生成热力图 海马、中额叶、杏仁核、颞叶 金祝新[53 ] AD/HC 3D-CNN(迁移学习) 90.9 输入添加遮挡块 内侧颞叶、海马体 Kwak[54 ] sMCI/pMCI DenseNet 73.90 输入添加遮挡块 海马、梭状回、颞下回、楔前叶 Venugopalan[55 ] AD/HC AE,CNN,随机森林 88 特征屏蔽 海马体、杏仁核 Shahamat[56 ] AD/HC 3D-CNN 85 遗传算法选取脑模板 左侧枕叶、左侧颞梭状皮层、右侧楔皮层、右额中回、右颞中回
对于AD影像学分类及其模型可解释性研究,未来有以下发展方向:(1)探索更多对AD分类有效的数据模态,以及更多基于多模态数据融合分类的方法;(2)针对各个医疗机构成像条件不同带来的数据差异,进行更多跨数据集的模型测试用于优化模型的泛化能力;(3)在AD与HC分类问题的基础上,改进不同类型MCI分类或多分类任务的准确度;(4)针对AD影像学分类相关任务的深度学习模型,进行更多的可解释性研究,并发现更多可靠的与AD疾病相关的脑区. ...
Functional brain network classification for Alzheimer’s disease detection with deep features and extreme learning machine
3
2020
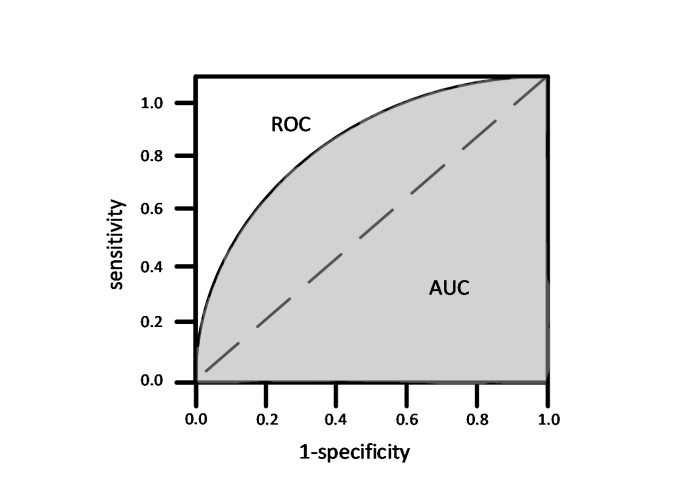
... 为了避免分类阈值设定导致准确率无法准确反应模型性能的问题,有些研究者[19 ] 会使用AUC对模型进行评估.AUC即接受者操作特征曲线(receiver operating characteristic curve,ROC)下方面积.为了介绍ROC,需要引入两个概念:敏感度(sensitivity),即真阳性样本在实际阳性样本中的占比;特异度(specificity),即真阴性样本在实际阴性样本中的占比.计算公式如(2)、(3)式所示: ...
... Parmar等[24 ] 从4D fMRI中选取连续时间序列的5个3D MRI作为3D CNN的输入,经过5个卷积层和3个全连接层,最后得到AD与HC分类准确率为94.58%的分类模型.Bi等[19 ] 基于AAL模板构建脑图谱,然后构建脑网络,通过RNN学习相邻位置特征,最后使用极限学习机(extreme learning machine,ELM)作为分类器,使AD与HC分类AUC达到了91.3%. ...
... Performance comparison of different models
Table 2 第一作者 分类任务 数据模态 数据集 测试方法 分类模型 准确率% 郁松[21 ] AD/HC sMRI 1015 HC 训练集60% 3D ResNet-101 97.425 Parmar[24 ] AD/HC fMRI 30 AD 训练集60% 3D CNN 94.58 Bi[19 ] AD/HC fMRI 118 AD 五折交叉验证 RNN,ELM 91.3 (AUC) Yi̇Ği̇T[20 ] AD/HC sMRI 训练集 测试集 / 2D CNN 83 Punjabi[33 ] AD/HC PET 共1299 / 3D CNN 85.15 Zhang[49 ] AD/HC sMRI 200 AD 五折交叉验证 3D ResAttNet 91.3 Qiu[52 ] AD/HC sMRI+性别+年龄+MMSE / 训练集60% FCN 96.8 Deng[28 ] AD/HC DTI+sMRI 98 AD 训练集60% CNN 90.00 Marzban[29 ] AD/HC DTI+sMRI 115 AD 十折交叉验证 CNN 93.5 Kang[30 ] EMCI/HC DTI+sMRI 50 HC 训练集80% VGG-16,LASSO 94.2 Kwak[54 ] AD/HC sMRI 110 AD 五折交叉验证 DenseNet 93.75 Ju[16 ] MCI/HC fMRI 91 MCI 十折交叉验证 AE 86.47 Baydargil[17 ] AD/MCI/HC PET 141 AD 训练集80% CAE 98.67 Guan[51 ] AD/HC sMRI 384 AD 训练集 90% ResNet、pABN 90.7 Li[34 ] AD/MCI/HC MRI 237 AD 训练集65% ResNet-200 83 Basaia[40 ] AD/HC sMRI 294 AD 训练集90% 3D FCN(迁移学习) 99.2 孔伶旭[36 ] EMCI/HC fMRI 32 HC 五折交叉验证 Mobilenet(迁移学习) 73.67 Bin[37 ] AD/HC sMRI 100 AD 五折交叉验证 Inception-v3 99.45 Massalimova[27 ] AD/MCI/HC DTI 训练集 测试集 / ResNet-18(迁移学习) 97 Mehmood[38 ] AD/HC sMRI 75 AD 训练集64% VGG-19(迁移学习) 95.33 Raju[50 ] 非常轻度痴呆/轻度痴呆/中度痴呆/HC(四分类) sMRI 1013非常轻度痴呆 334非常轻度痴呆 VGG-16(迁移学习) 99 Lian[39 ] AD/HC sMRI 数据集1 数据集2 两个数据集间 H-FCN(迁移学习) 90.3 Oh[18 ] AD/HC sMRI 198 AD 五折交叉验证 3D CNN,ICAE 88.6 金祝新[53 ] AD/MCI sMRI 267 AD 训练集90% 3D CNN(迁移学习) 94.6 曾安[42 ] AD/HC sMRI 137 AD 五折交叉验证 2D CNN(集成学习) 81 Bi[22 ] AD/HC sMRI 243 AD / PCANet,k -means 89.15 Choi[41 ] AD/HC sMRI 715 AD 训练集60% VGG-16,GoogLeNet,AlexNet(集成学习) 93.84 Venugopalan[55 ] AD/HC sMRI+电子病历+基因数据 共220 十折交叉验证 AE,CNN,随机森林 88 Zeng[46 ] sMCI/pMCI sMRI 92 AD 训练集70% PCA,DBN(多任务) 87.78 Spasov[43 ] sMCI/pMCI sMRI 192 AD 十折交叉验证 CNN(多任务) 86 Liu[48 ] AD/HC sMRI 97 AD 五折交叉验证 V-Net,DenseNet 88.9 Abuhmed[15 ] AD/MCI/HC PET+sMRI+神经心理学数据+神经病理学数据+认知评分 共1371 十折交叉验证 BiLSTM,随机森林 84.95
10.11938/cjmr20223013.T0003 表3 可解释性方法及AD相关脑区 ...
Applying deep learning models to structural MRI for stage prediction of Alzheimer’s disease
2
2020
... Yi̇Ği̇T等[20 ] 分别使用sMRI图像的轴向面、矢状面和冠状面作为CNN的输入,发现对于AD与HC分类任务而言,使用轴向面投影数据的分类准确率最高达到了83%;对于MCI与HC分类任务而言,使用矢状投影数据的分类准确率最高达到了82%.郁松等[21 ] 将3D sMRI作为输入,并使用3D ResNet-101网络,使AD与HC分类准确率达到了97.425%. ...
... Performance comparison of different models
Table 2 第一作者 分类任务 数据模态 数据集 测试方法 分类模型 准确率% 郁松[21 ] AD/HC sMRI 1015 HC 训练集60% 3D ResNet-101 97.425 Parmar[24 ] AD/HC fMRI 30 AD 训练集60% 3D CNN 94.58 Bi[19 ] AD/HC fMRI 118 AD 五折交叉验证 RNN,ELM 91.3 (AUC) Yi̇Ği̇T[20 ] AD/HC sMRI 训练集 测试集 / 2D CNN 83 Punjabi[33 ] AD/HC PET 共1299 / 3D CNN 85.15 Zhang[49 ] AD/HC sMRI 200 AD 五折交叉验证 3D ResAttNet 91.3 Qiu[52 ] AD/HC sMRI+性别+年龄+MMSE / 训练集60% FCN 96.8 Deng[28 ] AD/HC DTI+sMRI 98 AD 训练集60% CNN 90.00 Marzban[29 ] AD/HC DTI+sMRI 115 AD 十折交叉验证 CNN 93.5 Kang[30 ] EMCI/HC DTI+sMRI 50 HC 训练集80% VGG-16,LASSO 94.2 Kwak[54 ] AD/HC sMRI 110 AD 五折交叉验证 DenseNet 93.75 Ju[16 ] MCI/HC fMRI 91 MCI 十折交叉验证 AE 86.47 Baydargil[17 ] AD/MCI/HC PET 141 AD 训练集80% CAE 98.67 Guan[51 ] AD/HC sMRI 384 AD 训练集 90% ResNet、pABN 90.7 Li[34 ] AD/MCI/HC MRI 237 AD 训练集65% ResNet-200 83 Basaia[40 ] AD/HC sMRI 294 AD 训练集90% 3D FCN(迁移学习) 99.2 孔伶旭[36 ] EMCI/HC fMRI 32 HC 五折交叉验证 Mobilenet(迁移学习) 73.67 Bin[37 ] AD/HC sMRI 100 AD 五折交叉验证 Inception-v3 99.45 Massalimova[27 ] AD/MCI/HC DTI 训练集 测试集 / ResNet-18(迁移学习) 97 Mehmood[38 ] AD/HC sMRI 75 AD 训练集64% VGG-19(迁移学习) 95.33 Raju[50 ] 非常轻度痴呆/轻度痴呆/中度痴呆/HC(四分类) sMRI 1013非常轻度痴呆 334非常轻度痴呆 VGG-16(迁移学习) 99 Lian[39 ] AD/HC sMRI 数据集1 数据集2 两个数据集间 H-FCN(迁移学习) 90.3 Oh[18 ] AD/HC sMRI 198 AD 五折交叉验证 3D CNN,ICAE 88.6 金祝新[53 ] AD/MCI sMRI 267 AD 训练集90% 3D CNN(迁移学习) 94.6 曾安[42 ] AD/HC sMRI 137 AD 五折交叉验证 2D CNN(集成学习) 81 Bi[22 ] AD/HC sMRI 243 AD / PCANet,k -means 89.15 Choi[41 ] AD/HC sMRI 715 AD 训练集60% VGG-16,GoogLeNet,AlexNet(集成学习) 93.84 Venugopalan[55 ] AD/HC sMRI+电子病历+基因数据 共220 十折交叉验证 AE,CNN,随机森林 88 Zeng[46 ] sMCI/pMCI sMRI 92 AD 训练集70% PCA,DBN(多任务) 87.78 Spasov[43 ] sMCI/pMCI sMRI 192 AD 十折交叉验证 CNN(多任务) 86 Liu[48 ] AD/HC sMRI 97 AD 五折交叉验证 V-Net,DenseNet 88.9 Abuhmed[15 ] AD/MCI/HC PET+sMRI+神经心理学数据+神经病理学数据+认知评分 共1371 十折交叉验证 BiLSTM,随机森林 84.95
10.11938/cjmr20223013.T0003 表3 可解释性方法及AD相关脑区 ...
基于3D-ResNet的阿尔兹海默症分类算法研究
2
2020
... Yi̇Ği̇T等[20 ] 分别使用sMRI图像的轴向面、矢状面和冠状面作为CNN的输入,发现对于AD与HC分类任务而言,使用轴向面投影数据的分类准确率最高达到了83%;对于MCI与HC分类任务而言,使用矢状投影数据的分类准确率最高达到了82%.郁松等[21 ] 将3D sMRI作为输入,并使用3D ResNet-101网络,使AD与HC分类准确率达到了97.425%. ...
... Performance comparison of different models
Table 2 第一作者 分类任务 数据模态 数据集 测试方法 分类模型 准确率% 郁松[21 ] AD/HC sMRI 1015 HC 训练集60% 3D ResNet-101 97.425 Parmar[24 ] AD/HC fMRI 30 AD 训练集60% 3D CNN 94.58 Bi[19 ] AD/HC fMRI 118 AD 五折交叉验证 RNN,ELM 91.3 (AUC) Yi̇Ği̇T[20 ] AD/HC sMRI 训练集 测试集 / 2D CNN 83 Punjabi[33 ] AD/HC PET 共1299 / 3D CNN 85.15 Zhang[49 ] AD/HC sMRI 200 AD 五折交叉验证 3D ResAttNet 91.3 Qiu[52 ] AD/HC sMRI+性别+年龄+MMSE / 训练集60% FCN 96.8 Deng[28 ] AD/HC DTI+sMRI 98 AD 训练集60% CNN 90.00 Marzban[29 ] AD/HC DTI+sMRI 115 AD 十折交叉验证 CNN 93.5 Kang[30 ] EMCI/HC DTI+sMRI 50 HC 训练集80% VGG-16,LASSO 94.2 Kwak[54 ] AD/HC sMRI 110 AD 五折交叉验证 DenseNet 93.75 Ju[16 ] MCI/HC fMRI 91 MCI 十折交叉验证 AE 86.47 Baydargil[17 ] AD/MCI/HC PET 141 AD 训练集80% CAE 98.67 Guan[51 ] AD/HC sMRI 384 AD 训练集 90% ResNet、pABN 90.7 Li[34 ] AD/MCI/HC MRI 237 AD 训练集65% ResNet-200 83 Basaia[40 ] AD/HC sMRI 294 AD 训练集90% 3D FCN(迁移学习) 99.2 孔伶旭[36 ] EMCI/HC fMRI 32 HC 五折交叉验证 Mobilenet(迁移学习) 73.67 Bin[37 ] AD/HC sMRI 100 AD 五折交叉验证 Inception-v3 99.45 Massalimova[27 ] AD/MCI/HC DTI 训练集 测试集 / ResNet-18(迁移学习) 97 Mehmood[38 ] AD/HC sMRI 75 AD 训练集64% VGG-19(迁移学习) 95.33 Raju[50 ] 非常轻度痴呆/轻度痴呆/中度痴呆/HC(四分类) sMRI 1013非常轻度痴呆 334非常轻度痴呆 VGG-16(迁移学习) 99 Lian[39 ] AD/HC sMRI 数据集1 数据集2 两个数据集间 H-FCN(迁移学习) 90.3 Oh[18 ] AD/HC sMRI 198 AD 五折交叉验证 3D CNN,ICAE 88.6 金祝新[53 ] AD/MCI sMRI 267 AD 训练集90% 3D CNN(迁移学习) 94.6 曾安[42 ] AD/HC sMRI 137 AD 五折交叉验证 2D CNN(集成学习) 81 Bi[22 ] AD/HC sMRI 243 AD / PCANet,k -means 89.15 Choi[41 ] AD/HC sMRI 715 AD 训练集60% VGG-16,GoogLeNet,AlexNet(集成学习) 93.84 Venugopalan[55 ] AD/HC sMRI+电子病历+基因数据 共220 十折交叉验证 AE,CNN,随机森林 88 Zeng[46 ] sMCI/pMCI sMRI 92 AD 训练集70% PCA,DBN(多任务) 87.78 Spasov[43 ] sMCI/pMCI sMRI 192 AD 十折交叉验证 CNN(多任务) 86 Liu[48 ] AD/HC sMRI 97 AD 五折交叉验证 V-Net,DenseNet 88.9 Abuhmed[15 ] AD/MCI/HC PET+sMRI+神经心理学数据+神经病理学数据+认知评分 共1371 十折交叉验证 BiLSTM,随机森林 84.95
10.11938/cjmr20223013.T0003 表3 可解释性方法及AD相关脑区 ...
基于3D-ResNet的阿尔兹海默症分类算法研究
2
2020
... Yi̇Ği̇T等[20 ] 分别使用sMRI图像的轴向面、矢状面和冠状面作为CNN的输入,发现对于AD与HC分类任务而言,使用轴向面投影数据的分类准确率最高达到了83%;对于MCI与HC分类任务而言,使用矢状投影数据的分类准确率最高达到了82%.郁松等[21 ] 将3D sMRI作为输入,并使用3D ResNet-101网络,使AD与HC分类准确率达到了97.425%. ...
... Performance comparison of different models
Table 2 第一作者 分类任务 数据模态 数据集 测试方法 分类模型 准确率% 郁松[21 ] AD/HC sMRI 1015 HC 训练集60% 3D ResNet-101 97.425 Parmar[24 ] AD/HC fMRI 30 AD 训练集60% 3D CNN 94.58 Bi[19 ] AD/HC fMRI 118 AD 五折交叉验证 RNN,ELM 91.3 (AUC) Yi̇Ği̇T[20 ] AD/HC sMRI 训练集 测试集 / 2D CNN 83 Punjabi[33 ] AD/HC PET 共1299 / 3D CNN 85.15 Zhang[49 ] AD/HC sMRI 200 AD 五折交叉验证 3D ResAttNet 91.3 Qiu[52 ] AD/HC sMRI+性别+年龄+MMSE / 训练集60% FCN 96.8 Deng[28 ] AD/HC DTI+sMRI 98 AD 训练集60% CNN 90.00 Marzban[29 ] AD/HC DTI+sMRI 115 AD 十折交叉验证 CNN 93.5 Kang[30 ] EMCI/HC DTI+sMRI 50 HC 训练集80% VGG-16,LASSO 94.2 Kwak[54 ] AD/HC sMRI 110 AD 五折交叉验证 DenseNet 93.75 Ju[16 ] MCI/HC fMRI 91 MCI 十折交叉验证 AE 86.47 Baydargil[17 ] AD/MCI/HC PET 141 AD 训练集80% CAE 98.67 Guan[51 ] AD/HC sMRI 384 AD 训练集 90% ResNet、pABN 90.7 Li[34 ] AD/MCI/HC MRI 237 AD 训练集65% ResNet-200 83 Basaia[40 ] AD/HC sMRI 294 AD 训练集90% 3D FCN(迁移学习) 99.2 孔伶旭[36 ] EMCI/HC fMRI 32 HC 五折交叉验证 Mobilenet(迁移学习) 73.67 Bin[37 ] AD/HC sMRI 100 AD 五折交叉验证 Inception-v3 99.45 Massalimova[27 ] AD/MCI/HC DTI 训练集 测试集 / ResNet-18(迁移学习) 97 Mehmood[38 ] AD/HC sMRI 75 AD 训练集64% VGG-19(迁移学习) 95.33 Raju[50 ] 非常轻度痴呆/轻度痴呆/中度痴呆/HC(四分类) sMRI 1013非常轻度痴呆 334非常轻度痴呆 VGG-16(迁移学习) 99 Lian[39 ] AD/HC sMRI 数据集1 数据集2 两个数据集间 H-FCN(迁移学习) 90.3 Oh[18 ] AD/HC sMRI 198 AD 五折交叉验证 3D CNN,ICAE 88.6 金祝新[53 ] AD/MCI sMRI 267 AD 训练集90% 3D CNN(迁移学习) 94.6 曾安[42 ] AD/HC sMRI 137 AD 五折交叉验证 2D CNN(集成学习) 81 Bi[22 ] AD/HC sMRI 243 AD / PCANet,k -means 89.15 Choi[41 ] AD/HC sMRI 715 AD 训练集60% VGG-16,GoogLeNet,AlexNet(集成学习) 93.84 Venugopalan[55 ] AD/HC sMRI+电子病历+基因数据 共220 十折交叉验证 AE,CNN,随机森林 88 Zeng[46 ] sMCI/pMCI sMRI 92 AD 训练集70% PCA,DBN(多任务) 87.78 Spasov[43 ] sMCI/pMCI sMRI 192 AD 十折交叉验证 CNN(多任务) 86 Liu[48 ] AD/HC sMRI 97 AD 五折交叉验证 V-Net,DenseNet 88.9 Abuhmed[15 ] AD/MCI/HC PET+sMRI+神经心理学数据+神经病理学数据+认知评分 共1371 十折交叉验证 BiLSTM,随机森林 84.95
10.11938/cjmr20223013.T0003 表3 可解释性方法及AD相关脑区 ...
Computer aided Alzheimer's disease diagnosis by an unsupervised deep learning technology
2
2020
... 基于深度学习的图像分类任务需要大量的数据作为训练集,CNN作为一种监督学习模型,依赖于带有标签的图像,而带有标签的医学图像很难获得.为了解决这样的问题,Bi等[22 ] 提出了一种基于无监督学习的AD、MCI与HC分类模型,训练集使用不带标签的sMRI图像,结合主成分分析法(principal components analysis,PCA)与CNN对特征进行提取,使用k 均值聚类(k -means clustering algorithm,k -means)进行分类,最终实现AD与MCI的分类准确率为97.01%,AD、MCI与HC三分类准确率为91.25%. ...
... Performance comparison of different models
Table 2 第一作者 分类任务 数据模态 数据集 测试方法 分类模型 准确率% 郁松[21 ] AD/HC sMRI 1015 HC 训练集60% 3D ResNet-101 97.425 Parmar[24 ] AD/HC fMRI 30 AD 训练集60% 3D CNN 94.58 Bi[19 ] AD/HC fMRI 118 AD 五折交叉验证 RNN,ELM 91.3 (AUC) Yi̇Ği̇T[20 ] AD/HC sMRI 训练集 测试集 / 2D CNN 83 Punjabi[33 ] AD/HC PET 共1299 / 3D CNN 85.15 Zhang[49 ] AD/HC sMRI 200 AD 五折交叉验证 3D ResAttNet 91.3 Qiu[52 ] AD/HC sMRI+性别+年龄+MMSE / 训练集60% FCN 96.8 Deng[28 ] AD/HC DTI+sMRI 98 AD 训练集60% CNN 90.00 Marzban[29 ] AD/HC DTI+sMRI 115 AD 十折交叉验证 CNN 93.5 Kang[30 ] EMCI/HC DTI+sMRI 50 HC 训练集80% VGG-16,LASSO 94.2 Kwak[54 ] AD/HC sMRI 110 AD 五折交叉验证 DenseNet 93.75 Ju[16 ] MCI/HC fMRI 91 MCI 十折交叉验证 AE 86.47 Baydargil[17 ] AD/MCI/HC PET 141 AD 训练集80% CAE 98.67 Guan[51 ] AD/HC sMRI 384 AD 训练集 90% ResNet、pABN 90.7 Li[34 ] AD/MCI/HC MRI 237 AD 训练集65% ResNet-200 83 Basaia[40 ] AD/HC sMRI 294 AD 训练集90% 3D FCN(迁移学习) 99.2 孔伶旭[36 ] EMCI/HC fMRI 32 HC 五折交叉验证 Mobilenet(迁移学习) 73.67 Bin[37 ] AD/HC sMRI 100 AD 五折交叉验证 Inception-v3 99.45 Massalimova[27 ] AD/MCI/HC DTI 训练集 测试集 / ResNet-18(迁移学习) 97 Mehmood[38 ] AD/HC sMRI 75 AD 训练集64% VGG-19(迁移学习) 95.33 Raju[50 ] 非常轻度痴呆/轻度痴呆/中度痴呆/HC(四分类) sMRI 1013非常轻度痴呆 334非常轻度痴呆 VGG-16(迁移学习) 99 Lian[39 ] AD/HC sMRI 数据集1 数据集2 两个数据集间 H-FCN(迁移学习) 90.3 Oh[18 ] AD/HC sMRI 198 AD 五折交叉验证 3D CNN,ICAE 88.6 金祝新[53 ] AD/MCI sMRI 267 AD 训练集90% 3D CNN(迁移学习) 94.6 曾安[42 ] AD/HC sMRI 137 AD 五折交叉验证 2D CNN(集成学习) 81 Bi[22 ] AD/HC sMRI 243 AD / PCANet,k -means 89.15 Choi[41 ] AD/HC sMRI 715 AD 训练集60% VGG-16,GoogLeNet,AlexNet(集成学习) 93.84 Venugopalan[55 ] AD/HC sMRI+电子病历+基因数据 共220 十折交叉验证 AE,CNN,随机森林 88 Zeng[46 ] sMCI/pMCI sMRI 92 AD 训练集70% PCA,DBN(多任务) 87.78 Spasov[43 ] sMCI/pMCI sMRI 192 AD 十折交叉验证 CNN(多任务) 86 Liu[48 ] AD/HC sMRI 97 AD 五折交叉验证 V-Net,DenseNet 88.9 Abuhmed[15 ] AD/MCI/HC PET+sMRI+神经心理学数据+神经病理学数据+认知评分 共1371 十折交叉验证 BiLSTM,随机森林 84.95
10.11938/cjmr20223013.T0003 表3 可解释性方法及AD相关脑区 ...
Resting state fMRI and improved deep learning algorithm for earlier detection of Alzheimer’s disease
1
2020
... 也有很多研究者使用fMRI对AD分类问题进行研究.fMRI通过检测血氧水平依赖来估计大脑的活动情况,牺牲了空间分辨率,但提高了时间分辨率.fMRI可以用于研究针对特定任务的相关脑区,在心理学和认知科学中经常被使用.其中,静息态fMRI可以用于实现人脑功能区及功能网络的构建,同时也提供了重要的区域时序上的关系[23 ] .因此,对于影响认知功能的疾病,fMRI相比sMRI可以提供脑区功能的信息. ...
Deep learning of volumetric 3D CNN for fMRI in Alzheimer’s disease classification
2
2020
... Parmar等[24 ] 从4D fMRI中选取连续时间序列的5个3D MRI作为3D CNN的输入,经过5个卷积层和3个全连接层,最后得到AD与HC分类准确率为94.58%的分类模型.Bi等[19 ] 基于AAL模板构建脑图谱,然后构建脑网络,通过RNN学习相邻位置特征,最后使用极限学习机(extreme learning machine,ELM)作为分类器,使AD与HC分类AUC达到了91.3%. ...
... Performance comparison of different models
Table 2 第一作者 分类任务 数据模态 数据集 测试方法 分类模型 准确率% 郁松[21 ] AD/HC sMRI 1015 HC 训练集60% 3D ResNet-101 97.425 Parmar[24 ] AD/HC fMRI 30 AD 训练集60% 3D CNN 94.58 Bi[19 ] AD/HC fMRI 118 AD 五折交叉验证 RNN,ELM 91.3 (AUC) Yi̇Ği̇T[20 ] AD/HC sMRI 训练集 测试集 / 2D CNN 83 Punjabi[33 ] AD/HC PET 共1299 / 3D CNN 85.15 Zhang[49 ] AD/HC sMRI 200 AD 五折交叉验证 3D ResAttNet 91.3 Qiu[52 ] AD/HC sMRI+性别+年龄+MMSE / 训练集60% FCN 96.8 Deng[28 ] AD/HC DTI+sMRI 98 AD 训练集60% CNN 90.00 Marzban[29 ] AD/HC DTI+sMRI 115 AD 十折交叉验证 CNN 93.5 Kang[30 ] EMCI/HC DTI+sMRI 50 HC 训练集80% VGG-16,LASSO 94.2 Kwak[54 ] AD/HC sMRI 110 AD 五折交叉验证 DenseNet 93.75 Ju[16 ] MCI/HC fMRI 91 MCI 十折交叉验证 AE 86.47 Baydargil[17 ] AD/MCI/HC PET 141 AD 训练集80% CAE 98.67 Guan[51 ] AD/HC sMRI 384 AD 训练集 90% ResNet、pABN 90.7 Li[34 ] AD/MCI/HC MRI 237 AD 训练集65% ResNet-200 83 Basaia[40 ] AD/HC sMRI 294 AD 训练集90% 3D FCN(迁移学习) 99.2 孔伶旭[36 ] EMCI/HC fMRI 32 HC 五折交叉验证 Mobilenet(迁移学习) 73.67 Bin[37 ] AD/HC sMRI 100 AD 五折交叉验证 Inception-v3 99.45 Massalimova[27 ] AD/MCI/HC DTI 训练集 测试集 / ResNet-18(迁移学习) 97 Mehmood[38 ] AD/HC sMRI 75 AD 训练集64% VGG-19(迁移学习) 95.33 Raju[50 ] 非常轻度痴呆/轻度痴呆/中度痴呆/HC(四分类) sMRI 1013非常轻度痴呆 334非常轻度痴呆 VGG-16(迁移学习) 99 Lian[39 ] AD/HC sMRI 数据集1 数据集2 两个数据集间 H-FCN(迁移学习) 90.3 Oh[18 ] AD/HC sMRI 198 AD 五折交叉验证 3D CNN,ICAE 88.6 金祝新[53 ] AD/MCI sMRI 267 AD 训练集90% 3D CNN(迁移学习) 94.6 曾安[42 ] AD/HC sMRI 137 AD 五折交叉验证 2D CNN(集成学习) 81 Bi[22 ] AD/HC sMRI 243 AD / PCANet,k -means 89.15 Choi[41 ] AD/HC sMRI 715 AD 训练集60% VGG-16,GoogLeNet,AlexNet(集成学习) 93.84 Venugopalan[55 ] AD/HC sMRI+电子病历+基因数据 共220 十折交叉验证 AE,CNN,随机森林 88 Zeng[46 ] sMCI/pMCI sMRI 92 AD 训练集70% PCA,DBN(多任务) 87.78 Spasov[43 ] sMCI/pMCI sMRI 192 AD 十折交叉验证 CNN(多任务) 86 Liu[48 ] AD/HC sMRI 97 AD 五折交叉验证 V-Net,DenseNet 88.9 Abuhmed[15 ] AD/MCI/HC PET+sMRI+神经心理学数据+神经病理学数据+认知评分 共1371 十折交叉验证 BiLSTM,随机森林 84.95
10.11938/cjmr20223013.T0003 表3 可解释性方法及AD相关脑区 ...
Altered superficial white matter on tractography MRI in Alzheimer’s disease
1
2016
... 很多医学研究表明AD在脑影像上的特征表现为脑实质的萎缩,主要发生的区域为海马和颞叶.例如,Reginold等[25 ] 发现AD患者的颞叶出现很大程度的扩散异常,颞叶中浅表白质的平均扩散率(mean diffusivity,MD)、径向扩散系数(radical diffusion,RD)大幅上升.Bigham等[26 ] 发现MCI患者的右顶叶浅层白质的MD显著增加,AD患者的左边缘系统和左颞叶浅层白质的MD显著增加.而探测水分子扩散的DTI可以很好地对白质神经纤维束进行成像,直观地反应大脑白质的结构变化,所以有研究者使用DTI数据进行AD影像分类. ...
Identification of superficial white matter abnormalities in Alzheimer’s disease and mild cognitive impairment using diffusion tensor imaging
1
2020
... 很多医学研究表明AD在脑影像上的特征表现为脑实质的萎缩,主要发生的区域为海马和颞叶.例如,Reginold等[25 ] 发现AD患者的颞叶出现很大程度的扩散异常,颞叶中浅表白质的平均扩散率(mean diffusivity,MD)、径向扩散系数(radical diffusion,RD)大幅上升.Bigham等[26 ] 发现MCI患者的右顶叶浅层白质的MD显著增加,AD患者的左边缘系统和左颞叶浅层白质的MD显著增加.而探测水分子扩散的DTI可以很好地对白质神经纤维束进行成像,直观地反应大脑白质的结构变化,所以有研究者使用DTI数据进行AD影像分类. ...
Input agnostic deep learning for Alzheimer’s disease classification using multimodal MRI images
2
2021
... Massalimova等[27 ] 融合了MD扩散像、各向异性分数(fractional anisotropy,FA)扩散像和T 1 加权sMRI,使用ResNet-18作为网络主干,最终得到AD、MCI与HC的三分类准确率为97%.Deng等[28 ] 通过DTI图像计算得到中心度特征(degree centrality,DC),使用CNN作为分类器,使AD与HC分类准确率达到了90.00%.Marzban等[29 ] 将MD扩散像、FA扩散像和各向异性模式(mode of anisotropy,MO)扩散像作为2D CNN的输入,发现使用MD扩散像的AD与HC分类准确率最高达到88.9%,且融合MD扩散像和sMRI中的灰质图像可以取得更高的准确率93.5%.Kang等[30 ] 将sMRI、FA扩散像、MD扩散像通过三通道合并输入到VGG-16网络中进行特征提取,并使用最小绝对收缩和选择算子(Least absolute shrinkage and selection operator,LASSO)算法进行分类,最终使早期MCI(early mild cognitive impairment,EMCI)与HC分类准确率达到94.2%. ...
... Performance comparison of different models
Table 2 第一作者 分类任务 数据模态 数据集 测试方法 分类模型 准确率% 郁松[21 ] AD/HC sMRI 1015 HC 训练集60% 3D ResNet-101 97.425 Parmar[24 ] AD/HC fMRI 30 AD 训练集60% 3D CNN 94.58 Bi[19 ] AD/HC fMRI 118 AD 五折交叉验证 RNN,ELM 91.3 (AUC) Yi̇Ği̇T[20 ] AD/HC sMRI 训练集 测试集 / 2D CNN 83 Punjabi[33 ] AD/HC PET 共1299 / 3D CNN 85.15 Zhang[49 ] AD/HC sMRI 200 AD 五折交叉验证 3D ResAttNet 91.3 Qiu[52 ] AD/HC sMRI+性别+年龄+MMSE / 训练集60% FCN 96.8 Deng[28 ] AD/HC DTI+sMRI 98 AD 训练集60% CNN 90.00 Marzban[29 ] AD/HC DTI+sMRI 115 AD 十折交叉验证 CNN 93.5 Kang[30 ] EMCI/HC DTI+sMRI 50 HC 训练集80% VGG-16,LASSO 94.2 Kwak[54 ] AD/HC sMRI 110 AD 五折交叉验证 DenseNet 93.75 Ju[16 ] MCI/HC fMRI 91 MCI 十折交叉验证 AE 86.47 Baydargil[17 ] AD/MCI/HC PET 141 AD 训练集80% CAE 98.67 Guan[51 ] AD/HC sMRI 384 AD 训练集 90% ResNet、pABN 90.7 Li[34 ] AD/MCI/HC MRI 237 AD 训练集65% ResNet-200 83 Basaia[40 ] AD/HC sMRI 294 AD 训练集90% 3D FCN(迁移学习) 99.2 孔伶旭[36 ] EMCI/HC fMRI 32 HC 五折交叉验证 Mobilenet(迁移学习) 73.67 Bin[37 ] AD/HC sMRI 100 AD 五折交叉验证 Inception-v3 99.45 Massalimova[27 ] AD/MCI/HC DTI 训练集 测试集 / ResNet-18(迁移学习) 97 Mehmood[38 ] AD/HC sMRI 75 AD 训练集64% VGG-19(迁移学习) 95.33 Raju[50 ] 非常轻度痴呆/轻度痴呆/中度痴呆/HC(四分类) sMRI 1013非常轻度痴呆 334非常轻度痴呆 VGG-16(迁移学习) 99 Lian[39 ] AD/HC sMRI 数据集1 数据集2 两个数据集间 H-FCN(迁移学习) 90.3 Oh[18 ] AD/HC sMRI 198 AD 五折交叉验证 3D CNN,ICAE 88.6 金祝新[53 ] AD/MCI sMRI 267 AD 训练集90% 3D CNN(迁移学习) 94.6 曾安[42 ] AD/HC sMRI 137 AD 五折交叉验证 2D CNN(集成学习) 81 Bi[22 ] AD/HC sMRI 243 AD / PCANet,k -means 89.15 Choi[41 ] AD/HC sMRI 715 AD 训练集60% VGG-16,GoogLeNet,AlexNet(集成学习) 93.84 Venugopalan[55 ] AD/HC sMRI+电子病历+基因数据 共220 十折交叉验证 AE,CNN,随机森林 88 Zeng[46 ] sMCI/pMCI sMRI 92 AD 训练集70% PCA,DBN(多任务) 87.78 Spasov[43 ] sMCI/pMCI sMRI 192 AD 十折交叉验证 CNN(多任务) 86 Liu[48 ] AD/HC sMRI 97 AD 五折交叉验证 V-Net,DenseNet 88.9 Abuhmed[15 ] AD/MCI/HC PET+sMRI+神经心理学数据+神经病理学数据+认知评分 共1371 十折交叉验证 BiLSTM,随机森林 84.95
10.11938/cjmr20223013.T0003 表3 可解释性方法及AD相关脑区 ...
Hybrid diffusion tensor imaging feature-based AD classification
2
2021
... Massalimova等[27 ] 融合了MD扩散像、各向异性分数(fractional anisotropy,FA)扩散像和T 1 加权sMRI,使用ResNet-18作为网络主干,最终得到AD、MCI与HC的三分类准确率为97%.Deng等[28 ] 通过DTI图像计算得到中心度特征(degree centrality,DC),使用CNN作为分类器,使AD与HC分类准确率达到了90.00%.Marzban等[29 ] 将MD扩散像、FA扩散像和各向异性模式(mode of anisotropy,MO)扩散像作为2D CNN的输入,发现使用MD扩散像的AD与HC分类准确率最高达到88.9%,且融合MD扩散像和sMRI中的灰质图像可以取得更高的准确率93.5%.Kang等[30 ] 将sMRI、FA扩散像、MD扩散像通过三通道合并输入到VGG-16网络中进行特征提取,并使用最小绝对收缩和选择算子(Least absolute shrinkage and selection operator,LASSO)算法进行分类,最终使早期MCI(early mild cognitive impairment,EMCI)与HC分类准确率达到94.2%. ...
... Performance comparison of different models
Table 2 第一作者 分类任务 数据模态 数据集 测试方法 分类模型 准确率% 郁松[21 ] AD/HC sMRI 1015 HC 训练集60% 3D ResNet-101 97.425 Parmar[24 ] AD/HC fMRI 30 AD 训练集60% 3D CNN 94.58 Bi[19 ] AD/HC fMRI 118 AD 五折交叉验证 RNN,ELM 91.3 (AUC) Yi̇Ği̇T[20 ] AD/HC sMRI 训练集 测试集 / 2D CNN 83 Punjabi[33 ] AD/HC PET 共1299 / 3D CNN 85.15 Zhang[49 ] AD/HC sMRI 200 AD 五折交叉验证 3D ResAttNet 91.3 Qiu[52 ] AD/HC sMRI+性别+年龄+MMSE / 训练集60% FCN 96.8 Deng[28 ] AD/HC DTI+sMRI 98 AD 训练集60% CNN 90.00 Marzban[29 ] AD/HC DTI+sMRI 115 AD 十折交叉验证 CNN 93.5 Kang[30 ] EMCI/HC DTI+sMRI 50 HC 训练集80% VGG-16,LASSO 94.2 Kwak[54 ] AD/HC sMRI 110 AD 五折交叉验证 DenseNet 93.75 Ju[16 ] MCI/HC fMRI 91 MCI 十折交叉验证 AE 86.47 Baydargil[17 ] AD/MCI/HC PET 141 AD 训练集80% CAE 98.67 Guan[51 ] AD/HC sMRI 384 AD 训练集 90% ResNet、pABN 90.7 Li[34 ] AD/MCI/HC MRI 237 AD 训练集65% ResNet-200 83 Basaia[40 ] AD/HC sMRI 294 AD 训练集90% 3D FCN(迁移学习) 99.2 孔伶旭[36 ] EMCI/HC fMRI 32 HC 五折交叉验证 Mobilenet(迁移学习) 73.67 Bin[37 ] AD/HC sMRI 100 AD 五折交叉验证 Inception-v3 99.45 Massalimova[27 ] AD/MCI/HC DTI 训练集 测试集 / ResNet-18(迁移学习) 97 Mehmood[38 ] AD/HC sMRI 75 AD 训练集64% VGG-19(迁移学习) 95.33 Raju[50 ] 非常轻度痴呆/轻度痴呆/中度痴呆/HC(四分类) sMRI 1013非常轻度痴呆 334非常轻度痴呆 VGG-16(迁移学习) 99 Lian[39 ] AD/HC sMRI 数据集1 数据集2 两个数据集间 H-FCN(迁移学习) 90.3 Oh[18 ] AD/HC sMRI 198 AD 五折交叉验证 3D CNN,ICAE 88.6 金祝新[53 ] AD/MCI sMRI 267 AD 训练集90% 3D CNN(迁移学习) 94.6 曾安[42 ] AD/HC sMRI 137 AD 五折交叉验证 2D CNN(集成学习) 81 Bi[22 ] AD/HC sMRI 243 AD / PCANet,k -means 89.15 Choi[41 ] AD/HC sMRI 715 AD 训练集60% VGG-16,GoogLeNet,AlexNet(集成学习) 93.84 Venugopalan[55 ] AD/HC sMRI+电子病历+基因数据 共220 十折交叉验证 AE,CNN,随机森林 88 Zeng[46 ] sMCI/pMCI sMRI 92 AD 训练集70% PCA,DBN(多任务) 87.78 Spasov[43 ] sMCI/pMCI sMRI 192 AD 十折交叉验证 CNN(多任务) 86 Liu[48 ] AD/HC sMRI 97 AD 五折交叉验证 V-Net,DenseNet 88.9 Abuhmed[15 ] AD/MCI/HC PET+sMRI+神经心理学数据+神经病理学数据+认知评分 共1371 十折交叉验证 BiLSTM,随机森林 84.95
10.11938/cjmr20223013.T0003 表3 可解释性方法及AD相关脑区 ...
Alzheimer's disease diagnosis from diffusion tensor images using convolutional neural networks
2
2020
... Massalimova等[27 ] 融合了MD扩散像、各向异性分数(fractional anisotropy,FA)扩散像和T 1 加权sMRI,使用ResNet-18作为网络主干,最终得到AD、MCI与HC的三分类准确率为97%.Deng等[28 ] 通过DTI图像计算得到中心度特征(degree centrality,DC),使用CNN作为分类器,使AD与HC分类准确率达到了90.00%.Marzban等[29 ] 将MD扩散像、FA扩散像和各向异性模式(mode of anisotropy,MO)扩散像作为2D CNN的输入,发现使用MD扩散像的AD与HC分类准确率最高达到88.9%,且融合MD扩散像和sMRI中的灰质图像可以取得更高的准确率93.5%.Kang等[30 ] 将sMRI、FA扩散像、MD扩散像通过三通道合并输入到VGG-16网络中进行特征提取,并使用最小绝对收缩和选择算子(Least absolute shrinkage and selection operator,LASSO)算法进行分类,最终使早期MCI(early mild cognitive impairment,EMCI)与HC分类准确率达到94.2%. ...
... Performance comparison of different models
Table 2 第一作者 分类任务 数据模态 数据集 测试方法 分类模型 准确率% 郁松[21 ] AD/HC sMRI 1015 HC 训练集60% 3D ResNet-101 97.425 Parmar[24 ] AD/HC fMRI 30 AD 训练集60% 3D CNN 94.58 Bi[19 ] AD/HC fMRI 118 AD 五折交叉验证 RNN,ELM 91.3 (AUC) Yi̇Ği̇T[20 ] AD/HC sMRI 训练集 测试集 / 2D CNN 83 Punjabi[33 ] AD/HC PET 共1299 / 3D CNN 85.15 Zhang[49 ] AD/HC sMRI 200 AD 五折交叉验证 3D ResAttNet 91.3 Qiu[52 ] AD/HC sMRI+性别+年龄+MMSE / 训练集60% FCN 96.8 Deng[28 ] AD/HC DTI+sMRI 98 AD 训练集60% CNN 90.00 Marzban[29 ] AD/HC DTI+sMRI 115 AD 十折交叉验证 CNN 93.5 Kang[30 ] EMCI/HC DTI+sMRI 50 HC 训练集80% VGG-16,LASSO 94.2 Kwak[54 ] AD/HC sMRI 110 AD 五折交叉验证 DenseNet 93.75 Ju[16 ] MCI/HC fMRI 91 MCI 十折交叉验证 AE 86.47 Baydargil[17 ] AD/MCI/HC PET 141 AD 训练集80% CAE 98.67 Guan[51 ] AD/HC sMRI 384 AD 训练集 90% ResNet、pABN 90.7 Li[34 ] AD/MCI/HC MRI 237 AD 训练集65% ResNet-200 83 Basaia[40 ] AD/HC sMRI 294 AD 训练集90% 3D FCN(迁移学习) 99.2 孔伶旭[36 ] EMCI/HC fMRI 32 HC 五折交叉验证 Mobilenet(迁移学习) 73.67 Bin[37 ] AD/HC sMRI 100 AD 五折交叉验证 Inception-v3 99.45 Massalimova[27 ] AD/MCI/HC DTI 训练集 测试集 / ResNet-18(迁移学习) 97 Mehmood[38 ] AD/HC sMRI 75 AD 训练集64% VGG-19(迁移学习) 95.33 Raju[50 ] 非常轻度痴呆/轻度痴呆/中度痴呆/HC(四分类) sMRI 1013非常轻度痴呆 334非常轻度痴呆 VGG-16(迁移学习) 99 Lian[39 ] AD/HC sMRI 数据集1 数据集2 两个数据集间 H-FCN(迁移学习) 90.3 Oh[18 ] AD/HC sMRI 198 AD 五折交叉验证 3D CNN,ICAE 88.6 金祝新[53 ] AD/MCI sMRI 267 AD 训练集90% 3D CNN(迁移学习) 94.6 曾安[42 ] AD/HC sMRI 137 AD 五折交叉验证 2D CNN(集成学习) 81 Bi[22 ] AD/HC sMRI 243 AD / PCANet,k -means 89.15 Choi[41 ] AD/HC sMRI 715 AD 训练集60% VGG-16,GoogLeNet,AlexNet(集成学习) 93.84 Venugopalan[55 ] AD/HC sMRI+电子病历+基因数据 共220 十折交叉验证 AE,CNN,随机森林 88 Zeng[46 ] sMCI/pMCI sMRI 92 AD 训练集70% PCA,DBN(多任务) 87.78 Spasov[43 ] sMCI/pMCI sMRI 192 AD 十折交叉验证 CNN(多任务) 86 Liu[48 ] AD/HC sMRI 97 AD 五折交叉验证 V-Net,DenseNet 88.9 Abuhmed[15 ] AD/MCI/HC PET+sMRI+神经心理学数据+神经病理学数据+认知评分 共1371 十折交叉验证 BiLSTM,随机森林 84.95
10.11938/cjmr20223013.T0003 表3 可解释性方法及AD相关脑区 ...
Identifying early mild cognitive impairment by multi-modality MRI-based deep learning
2
2020
... Massalimova等[27 ] 融合了MD扩散像、各向异性分数(fractional anisotropy,FA)扩散像和T 1 加权sMRI,使用ResNet-18作为网络主干,最终得到AD、MCI与HC的三分类准确率为97%.Deng等[28 ] 通过DTI图像计算得到中心度特征(degree centrality,DC),使用CNN作为分类器,使AD与HC分类准确率达到了90.00%.Marzban等[29 ] 将MD扩散像、FA扩散像和各向异性模式(mode of anisotropy,MO)扩散像作为2D CNN的输入,发现使用MD扩散像的AD与HC分类准确率最高达到88.9%,且融合MD扩散像和sMRI中的灰质图像可以取得更高的准确率93.5%.Kang等[30 ] 将sMRI、FA扩散像、MD扩散像通过三通道合并输入到VGG-16网络中进行特征提取,并使用最小绝对收缩和选择算子(Least absolute shrinkage and selection operator,LASSO)算法进行分类,最终使早期MCI(early mild cognitive impairment,EMCI)与HC分类准确率达到94.2%. ...
... Performance comparison of different models
Table 2 第一作者 分类任务 数据模态 数据集 测试方法 分类模型 准确率% 郁松[21 ] AD/HC sMRI 1015 HC 训练集60% 3D ResNet-101 97.425 Parmar[24 ] AD/HC fMRI 30 AD 训练集60% 3D CNN 94.58 Bi[19 ] AD/HC fMRI 118 AD 五折交叉验证 RNN,ELM 91.3 (AUC) Yi̇Ği̇T[20 ] AD/HC sMRI 训练集 测试集 / 2D CNN 83 Punjabi[33 ] AD/HC PET 共1299 / 3D CNN 85.15 Zhang[49 ] AD/HC sMRI 200 AD 五折交叉验证 3D ResAttNet 91.3 Qiu[52 ] AD/HC sMRI+性别+年龄+MMSE / 训练集60% FCN 96.8 Deng[28 ] AD/HC DTI+sMRI 98 AD 训练集60% CNN 90.00 Marzban[29 ] AD/HC DTI+sMRI 115 AD 十折交叉验证 CNN 93.5 Kang[30 ] EMCI/HC DTI+sMRI 50 HC 训练集80% VGG-16,LASSO 94.2 Kwak[54 ] AD/HC sMRI 110 AD 五折交叉验证 DenseNet 93.75 Ju[16 ] MCI/HC fMRI 91 MCI 十折交叉验证 AE 86.47 Baydargil[17 ] AD/MCI/HC PET 141 AD 训练集80% CAE 98.67 Guan[51 ] AD/HC sMRI 384 AD 训练集 90% ResNet、pABN 90.7 Li[34 ] AD/MCI/HC MRI 237 AD 训练集65% ResNet-200 83 Basaia[40 ] AD/HC sMRI 294 AD 训练集90% 3D FCN(迁移学习) 99.2 孔伶旭[36 ] EMCI/HC fMRI 32 HC 五折交叉验证 Mobilenet(迁移学习) 73.67 Bin[37 ] AD/HC sMRI 100 AD 五折交叉验证 Inception-v3 99.45 Massalimova[27 ] AD/MCI/HC DTI 训练集 测试集 / ResNet-18(迁移学习) 97 Mehmood[38 ] AD/HC sMRI 75 AD 训练集64% VGG-19(迁移学习) 95.33 Raju[50 ] 非常轻度痴呆/轻度痴呆/中度痴呆/HC(四分类) sMRI 1013非常轻度痴呆 334非常轻度痴呆 VGG-16(迁移学习) 99 Lian[39 ] AD/HC sMRI 数据集1 数据集2 两个数据集间 H-FCN(迁移学习) 90.3 Oh[18 ] AD/HC sMRI 198 AD 五折交叉验证 3D CNN,ICAE 88.6 金祝新[53 ] AD/MCI sMRI 267 AD 训练集90% 3D CNN(迁移学习) 94.6 曾安[42 ] AD/HC sMRI 137 AD 五折交叉验证 2D CNN(集成学习) 81 Bi[22 ] AD/HC sMRI 243 AD / PCANet,k -means 89.15 Choi[41 ] AD/HC sMRI 715 AD 训练集60% VGG-16,GoogLeNet,AlexNet(集成学习) 93.84 Venugopalan[55 ] AD/HC sMRI+电子病历+基因数据 共220 十折交叉验证 AE,CNN,随机森林 88 Zeng[46 ] sMCI/pMCI sMRI 92 AD 训练集70% PCA,DBN(多任务) 87.78 Spasov[43 ] sMCI/pMCI sMRI 192 AD 十折交叉验证 CNN(多任务) 86 Liu[48 ] AD/HC sMRI 97 AD 五折交叉验证 V-Net,DenseNet 88.9 Abuhmed[15 ] AD/MCI/HC PET+sMRI+神经心理学数据+神经病理学数据+认知评分 共1371 十折交叉验证 BiLSTM,随机森林 84.95
10.11938/cjmr20223013.T0003 表3 可解释性方法及AD相关脑区 ...
Integrated positron emission tomography/magnetic resonance imaging in clinical diagnosis of Alzheimer’s disease
1
2021
... PET是核医学中比较先进的成像技术,由于AD的一个显著特征是大脑中淀粉样蛋白斑块的累积,通过淀粉样蛋白的PET成像,医生可以检测AD患者的脑斑.有研究[31 ] 表明淀粉样蛋白PET图像的引入对于AD疾病的诊断产生了很大的影响.对于PET脑部扫描显示存在显著淀粉样沉积物的患者,分别有82%的MCI患者与91%的痴呆患者被临床医生建议服用针对AD的药物;而在进行PET扫描之前,仅有40%的MCI患者与63%的痴呆症患者服用针对AD的药物[32 ] .可见淀粉样蛋白阳性与AD之间高度相关,在基于PET数据的深度学习AD影像分类中也可以得到印证. ...
Association of amyloid positron emission tomography with subsequent change in clinical management among medicare beneficiaries with mild cognitive impairment or dementia
1
2019
... PET是核医学中比较先进的成像技术,由于AD的一个显著特征是大脑中淀粉样蛋白斑块的累积,通过淀粉样蛋白的PET成像,医生可以检测AD患者的脑斑.有研究[31 ] 表明淀粉样蛋白PET图像的引入对于AD疾病的诊断产生了很大的影响.对于PET脑部扫描显示存在显著淀粉样沉积物的患者,分别有82%的MCI患者与91%的痴呆患者被临床医生建议服用针对AD的药物;而在进行PET扫描之前,仅有40%的MCI患者与63%的痴呆症患者服用针对AD的药物[32 ] .可见淀粉样蛋白阳性与AD之间高度相关,在基于PET数据的深度学习AD影像分类中也可以得到印证. ...
Neuroimaging modality fusion in Alzheimer’s classification using convolutional neural networks
2
2019
... Punjabi等[33 ] 详细比较了sMRI图像与AV-45淀粉样蛋白PET图像对AD与HC分类效能的影响.文中采用1 299份sMRI数据和585份AV-45淀粉样蛋白PET数据,并且分别使用全部的sMRI数据、全部的PET数据、和PET数据量相同的sMRI数据以及PET与sMRI多模态数据对CNN进行训练.结果显示在AD和HC分类中,仅使用sMRI大约一半数量的PET数据就达到了85.15%的准确率,和使用全部sMRI达到的准确率87.49%相当,进一步使用PET与sMRI多模态数据则可以将准确率提升至92.34%.而基于PET实验中的假阳性样本均伴有淀粉样蛋白水平升高,印证了淀粉样蛋白水平升高与AD疾病高度相关.也可见PET数据相比sMRI数据在AD影像学分类上具有更高的效能,而两种数据的结合可以获得更高的分类性能. ...
... Performance comparison of different models
Table 2 第一作者 分类任务 数据模态 数据集 测试方法 分类模型 准确率% 郁松[21 ] AD/HC sMRI 1015 HC 训练集60% 3D ResNet-101 97.425 Parmar[24 ] AD/HC fMRI 30 AD 训练集60% 3D CNN 94.58 Bi[19 ] AD/HC fMRI 118 AD 五折交叉验证 RNN,ELM 91.3 (AUC) Yi̇Ği̇T[20 ] AD/HC sMRI 训练集 测试集 / 2D CNN 83 Punjabi[33 ] AD/HC PET 共1299 / 3D CNN 85.15 Zhang[49 ] AD/HC sMRI 200 AD 五折交叉验证 3D ResAttNet 91.3 Qiu[52 ] AD/HC sMRI+性别+年龄+MMSE / 训练集60% FCN 96.8 Deng[28 ] AD/HC DTI+sMRI 98 AD 训练集60% CNN 90.00 Marzban[29 ] AD/HC DTI+sMRI 115 AD 十折交叉验证 CNN 93.5 Kang[30 ] EMCI/HC DTI+sMRI 50 HC 训练集80% VGG-16,LASSO 94.2 Kwak[54 ] AD/HC sMRI 110 AD 五折交叉验证 DenseNet 93.75 Ju[16 ] MCI/HC fMRI 91 MCI 十折交叉验证 AE 86.47 Baydargil[17 ] AD/MCI/HC PET 141 AD 训练集80% CAE 98.67 Guan[51 ] AD/HC sMRI 384 AD 训练集 90% ResNet、pABN 90.7 Li[34 ] AD/MCI/HC MRI 237 AD 训练集65% ResNet-200 83 Basaia[40 ] AD/HC sMRI 294 AD 训练集90% 3D FCN(迁移学习) 99.2 孔伶旭[36 ] EMCI/HC fMRI 32 HC 五折交叉验证 Mobilenet(迁移学习) 73.67 Bin[37 ] AD/HC sMRI 100 AD 五折交叉验证 Inception-v3 99.45 Massalimova[27 ] AD/MCI/HC DTI 训练集 测试集 / ResNet-18(迁移学习) 97 Mehmood[38 ] AD/HC sMRI 75 AD 训练集64% VGG-19(迁移学习) 95.33 Raju[50 ] 非常轻度痴呆/轻度痴呆/中度痴呆/HC(四分类) sMRI 1013非常轻度痴呆 334非常轻度痴呆 VGG-16(迁移学习) 99 Lian[39 ] AD/HC sMRI 数据集1 数据集2 两个数据集间 H-FCN(迁移学习) 90.3 Oh[18 ] AD/HC sMRI 198 AD 五折交叉验证 3D CNN,ICAE 88.6 金祝新[53 ] AD/MCI sMRI 267 AD 训练集90% 3D CNN(迁移学习) 94.6 曾安[42 ] AD/HC sMRI 137 AD 五折交叉验证 2D CNN(集成学习) 81 Bi[22 ] AD/HC sMRI 243 AD / PCANet,k -means 89.15 Choi[41 ] AD/HC sMRI 715 AD 训练集60% VGG-16,GoogLeNet,AlexNet(集成学习) 93.84 Venugopalan[55 ] AD/HC sMRI+电子病历+基因数据 共220 十折交叉验证 AE,CNN,随机森林 88 Zeng[46 ] sMCI/pMCI sMRI 92 AD 训练集70% PCA,DBN(多任务) 87.78 Spasov[43 ] sMCI/pMCI sMRI 192 AD 十折交叉验证 CNN(多任务) 86 Liu[48 ] AD/HC sMRI 97 AD 五折交叉验证 V-Net,DenseNet 88.9 Abuhmed[15 ] AD/MCI/HC PET+sMRI+神经心理学数据+神经病理学数据+认知评分 共1371 十折交叉验证 BiLSTM,随机森林 84.95
10.11938/cjmr20223013.T0003 表3 可解释性方法及AD相关脑区 ...
Alzheimer’s disease classification model based on MED-3D transfer learning
2
2021
... Li等[34 ] 使用由文献[35 ]提供的3DSeg-8数据集作为预训练的源数据集,将权重迁移学习至ResNet-200,使AD、MCI和HC三分类准确率达到83%,而不使用迁移学习的准确率仅为76%.孔伶旭等[36 ] 使用ImageNet数据集迁移学习到Mobilenet网络的瓶颈层,然后将fMRI数据提取出来的ROI时间序列输入到网络中对网络顶层进行训练,最终网络对于EMCI与HC的分类准确率达到了73.67%.Bin等[37 ] 分别构建了平衡与不平衡数据集,同时在2D CNN、基于ImageNet迁移学习的Xception和Inception-v3网络中进行训练,最终发现带有迁移学习策略的Inception-v3在平衡数据集上性能最高,AD与HC分类准确率达到了99.45%. ...
... Performance comparison of different models
Table 2 第一作者 分类任务 数据模态 数据集 测试方法 分类模型 准确率% 郁松[21 ] AD/HC sMRI 1015 HC 训练集60% 3D ResNet-101 97.425 Parmar[24 ] AD/HC fMRI 30 AD 训练集60% 3D CNN 94.58 Bi[19 ] AD/HC fMRI 118 AD 五折交叉验证 RNN,ELM 91.3 (AUC) Yi̇Ği̇T[20 ] AD/HC sMRI 训练集 测试集 / 2D CNN 83 Punjabi[33 ] AD/HC PET 共1299 / 3D CNN 85.15 Zhang[49 ] AD/HC sMRI 200 AD 五折交叉验证 3D ResAttNet 91.3 Qiu[52 ] AD/HC sMRI+性别+年龄+MMSE / 训练集60% FCN 96.8 Deng[28 ] AD/HC DTI+sMRI 98 AD 训练集60% CNN 90.00 Marzban[29 ] AD/HC DTI+sMRI 115 AD 十折交叉验证 CNN 93.5 Kang[30 ] EMCI/HC DTI+sMRI 50 HC 训练集80% VGG-16,LASSO 94.2 Kwak[54 ] AD/HC sMRI 110 AD 五折交叉验证 DenseNet 93.75 Ju[16 ] MCI/HC fMRI 91 MCI 十折交叉验证 AE 86.47 Baydargil[17 ] AD/MCI/HC PET 141 AD 训练集80% CAE 98.67 Guan[51 ] AD/HC sMRI 384 AD 训练集 90% ResNet、pABN 90.7 Li[34 ] AD/MCI/HC MRI 237 AD 训练集65% ResNet-200 83 Basaia[40 ] AD/HC sMRI 294 AD 训练集90% 3D FCN(迁移学习) 99.2 孔伶旭[36 ] EMCI/HC fMRI 32 HC 五折交叉验证 Mobilenet(迁移学习) 73.67 Bin[37 ] AD/HC sMRI 100 AD 五折交叉验证 Inception-v3 99.45 Massalimova[27 ] AD/MCI/HC DTI 训练集 测试集 / ResNet-18(迁移学习) 97 Mehmood[38 ] AD/HC sMRI 75 AD 训练集64% VGG-19(迁移学习) 95.33 Raju[50 ] 非常轻度痴呆/轻度痴呆/中度痴呆/HC(四分类) sMRI 1013非常轻度痴呆 334非常轻度痴呆 VGG-16(迁移学习) 99 Lian[39 ] AD/HC sMRI 数据集1 数据集2 两个数据集间 H-FCN(迁移学习) 90.3 Oh[18 ] AD/HC sMRI 198 AD 五折交叉验证 3D CNN,ICAE 88.6 金祝新[53 ] AD/MCI sMRI 267 AD 训练集90% 3D CNN(迁移学习) 94.6 曾安[42 ] AD/HC sMRI 137 AD 五折交叉验证 2D CNN(集成学习) 81 Bi[22 ] AD/HC sMRI 243 AD / PCANet,k -means 89.15 Choi[41 ] AD/HC sMRI 715 AD 训练集60% VGG-16,GoogLeNet,AlexNet(集成学习) 93.84 Venugopalan[55 ] AD/HC sMRI+电子病历+基因数据 共220 十折交叉验证 AE,CNN,随机森林 88 Zeng[46 ] sMCI/pMCI sMRI 92 AD 训练集70% PCA,DBN(多任务) 87.78 Spasov[43 ] sMCI/pMCI sMRI 192 AD 十折交叉验证 CNN(多任务) 86 Liu[48 ] AD/HC sMRI 97 AD 五折交叉验证 V-Net,DenseNet 88.9 Abuhmed[15 ] AD/MCI/HC PET+sMRI+神经心理学数据+神经病理学数据+认知评分 共1371 十折交叉验证 BiLSTM,随机森林 84.95
10.11938/cjmr20223013.T0003 表3 可解释性方法及AD相关脑区 ...
Med3d: Transfer learning for 3d medical image analysis
1
... Li等[34 ] 使用由文献[35 ]提供的3DSeg-8数据集作为预训练的源数据集,将权重迁移学习至ResNet-200,使AD、MCI和HC三分类准确率达到83%,而不使用迁移学习的准确率仅为76%.孔伶旭等[36 ] 使用ImageNet数据集迁移学习到Mobilenet网络的瓶颈层,然后将fMRI数据提取出来的ROI时间序列输入到网络中对网络顶层进行训练,最终网络对于EMCI与HC的分类准确率达到了73.67%.Bin等[37 ] 分别构建了平衡与不平衡数据集,同时在2D CNN、基于ImageNet迁移学习的Xception和Inception-v3网络中进行训练,最终发现带有迁移学习策略的Inception-v3在平衡数据集上性能最高,AD与HC分类准确率达到了99.45%. ...
迁移学习特征提取的rs-fMRI早期轻度认知障碍分类
2
2021
... Li等[34 ] 使用由文献[35 ]提供的3DSeg-8数据集作为预训练的源数据集,将权重迁移学习至ResNet-200,使AD、MCI和HC三分类准确率达到83%,而不使用迁移学习的准确率仅为76%.孔伶旭等[36 ] 使用ImageNet数据集迁移学习到Mobilenet网络的瓶颈层,然后将fMRI数据提取出来的ROI时间序列输入到网络中对网络顶层进行训练,最终网络对于EMCI与HC的分类准确率达到了73.67%.Bin等[37 ] 分别构建了平衡与不平衡数据集,同时在2D CNN、基于ImageNet迁移学习的Xception和Inception-v3网络中进行训练,最终发现带有迁移学习策略的Inception-v3在平衡数据集上性能最高,AD与HC分类准确率达到了99.45%. ...
... Performance comparison of different models
Table 2 第一作者 分类任务 数据模态 数据集 测试方法 分类模型 准确率% 郁松[21 ] AD/HC sMRI 1015 HC 训练集60% 3D ResNet-101 97.425 Parmar[24 ] AD/HC fMRI 30 AD 训练集60% 3D CNN 94.58 Bi[19 ] AD/HC fMRI 118 AD 五折交叉验证 RNN,ELM 91.3 (AUC) Yi̇Ği̇T[20 ] AD/HC sMRI 训练集 测试集 / 2D CNN 83 Punjabi[33 ] AD/HC PET 共1299 / 3D CNN 85.15 Zhang[49 ] AD/HC sMRI 200 AD 五折交叉验证 3D ResAttNet 91.3 Qiu[52 ] AD/HC sMRI+性别+年龄+MMSE / 训练集60% FCN 96.8 Deng[28 ] AD/HC DTI+sMRI 98 AD 训练集60% CNN 90.00 Marzban[29 ] AD/HC DTI+sMRI 115 AD 十折交叉验证 CNN 93.5 Kang[30 ] EMCI/HC DTI+sMRI 50 HC 训练集80% VGG-16,LASSO 94.2 Kwak[54 ] AD/HC sMRI 110 AD 五折交叉验证 DenseNet 93.75 Ju[16 ] MCI/HC fMRI 91 MCI 十折交叉验证 AE 86.47 Baydargil[17 ] AD/MCI/HC PET 141 AD 训练集80% CAE 98.67 Guan[51 ] AD/HC sMRI 384 AD 训练集 90% ResNet、pABN 90.7 Li[34 ] AD/MCI/HC MRI 237 AD 训练集65% ResNet-200 83 Basaia[40 ] AD/HC sMRI 294 AD 训练集90% 3D FCN(迁移学习) 99.2 孔伶旭[36 ] EMCI/HC fMRI 32 HC 五折交叉验证 Mobilenet(迁移学习) 73.67 Bin[37 ] AD/HC sMRI 100 AD 五折交叉验证 Inception-v3 99.45 Massalimova[27 ] AD/MCI/HC DTI 训练集 测试集 / ResNet-18(迁移学习) 97 Mehmood[38 ] AD/HC sMRI 75 AD 训练集64% VGG-19(迁移学习) 95.33 Raju[50 ] 非常轻度痴呆/轻度痴呆/中度痴呆/HC(四分类) sMRI 1013非常轻度痴呆 334非常轻度痴呆 VGG-16(迁移学习) 99 Lian[39 ] AD/HC sMRI 数据集1 数据集2 两个数据集间 H-FCN(迁移学习) 90.3 Oh[18 ] AD/HC sMRI 198 AD 五折交叉验证 3D CNN,ICAE 88.6 金祝新[53 ] AD/MCI sMRI 267 AD 训练集90% 3D CNN(迁移学习) 94.6 曾安[42 ] AD/HC sMRI 137 AD 五折交叉验证 2D CNN(集成学习) 81 Bi[22 ] AD/HC sMRI 243 AD / PCANet,k -means 89.15 Choi[41 ] AD/HC sMRI 715 AD 训练集60% VGG-16,GoogLeNet,AlexNet(集成学习) 93.84 Venugopalan[55 ] AD/HC sMRI+电子病历+基因数据 共220 十折交叉验证 AE,CNN,随机森林 88 Zeng[46 ] sMCI/pMCI sMRI 92 AD 训练集70% PCA,DBN(多任务) 87.78 Spasov[43 ] sMCI/pMCI sMRI 192 AD 十折交叉验证 CNN(多任务) 86 Liu[48 ] AD/HC sMRI 97 AD 五折交叉验证 V-Net,DenseNet 88.9 Abuhmed[15 ] AD/MCI/HC PET+sMRI+神经心理学数据+神经病理学数据+认知评分 共1371 十折交叉验证 BiLSTM,随机森林 84.95
10.11938/cjmr20223013.T0003 表3 可解释性方法及AD相关脑区 ...
迁移学习特征提取的rs-fMRI早期轻度认知障碍分类
2
2021
... Li等[34 ] 使用由文献[35 ]提供的3DSeg-8数据集作为预训练的源数据集,将权重迁移学习至ResNet-200,使AD、MCI和HC三分类准确率达到83%,而不使用迁移学习的准确率仅为76%.孔伶旭等[36 ] 使用ImageNet数据集迁移学习到Mobilenet网络的瓶颈层,然后将fMRI数据提取出来的ROI时间序列输入到网络中对网络顶层进行训练,最终网络对于EMCI与HC的分类准确率达到了73.67%.Bin等[37 ] 分别构建了平衡与不平衡数据集,同时在2D CNN、基于ImageNet迁移学习的Xception和Inception-v3网络中进行训练,最终发现带有迁移学习策略的Inception-v3在平衡数据集上性能最高,AD与HC分类准确率达到了99.45%. ...
... Performance comparison of different models
Table 2 第一作者 分类任务 数据模态 数据集 测试方法 分类模型 准确率% 郁松[21 ] AD/HC sMRI 1015 HC 训练集60% 3D ResNet-101 97.425 Parmar[24 ] AD/HC fMRI 30 AD 训练集60% 3D CNN 94.58 Bi[19 ] AD/HC fMRI 118 AD 五折交叉验证 RNN,ELM 91.3 (AUC) Yi̇Ği̇T[20 ] AD/HC sMRI 训练集 测试集 / 2D CNN 83 Punjabi[33 ] AD/HC PET 共1299 / 3D CNN 85.15 Zhang[49 ] AD/HC sMRI 200 AD 五折交叉验证 3D ResAttNet 91.3 Qiu[52 ] AD/HC sMRI+性别+年龄+MMSE / 训练集60% FCN 96.8 Deng[28 ] AD/HC DTI+sMRI 98 AD 训练集60% CNN 90.00 Marzban[29 ] AD/HC DTI+sMRI 115 AD 十折交叉验证 CNN 93.5 Kang[30 ] EMCI/HC DTI+sMRI 50 HC 训练集80% VGG-16,LASSO 94.2 Kwak[54 ] AD/HC sMRI 110 AD 五折交叉验证 DenseNet 93.75 Ju[16 ] MCI/HC fMRI 91 MCI 十折交叉验证 AE 86.47 Baydargil[17 ] AD/MCI/HC PET 141 AD 训练集80% CAE 98.67 Guan[51 ] AD/HC sMRI 384 AD 训练集 90% ResNet、pABN 90.7 Li[34 ] AD/MCI/HC MRI 237 AD 训练集65% ResNet-200 83 Basaia[40 ] AD/HC sMRI 294 AD 训练集90% 3D FCN(迁移学习) 99.2 孔伶旭[36 ] EMCI/HC fMRI 32 HC 五折交叉验证 Mobilenet(迁移学习) 73.67 Bin[37 ] AD/HC sMRI 100 AD 五折交叉验证 Inception-v3 99.45 Massalimova[27 ] AD/MCI/HC DTI 训练集 测试集 / ResNet-18(迁移学习) 97 Mehmood[38 ] AD/HC sMRI 75 AD 训练集64% VGG-19(迁移学习) 95.33 Raju[50 ] 非常轻度痴呆/轻度痴呆/中度痴呆/HC(四分类) sMRI 1013非常轻度痴呆 334非常轻度痴呆 VGG-16(迁移学习) 99 Lian[39 ] AD/HC sMRI 数据集1 数据集2 两个数据集间 H-FCN(迁移学习) 90.3 Oh[18 ] AD/HC sMRI 198 AD 五折交叉验证 3D CNN,ICAE 88.6 金祝新[53 ] AD/MCI sMRI 267 AD 训练集90% 3D CNN(迁移学习) 94.6 曾安[42 ] AD/HC sMRI 137 AD 五折交叉验证 2D CNN(集成学习) 81 Bi[22 ] AD/HC sMRI 243 AD / PCANet,k -means 89.15 Choi[41 ] AD/HC sMRI 715 AD 训练集60% VGG-16,GoogLeNet,AlexNet(集成学习) 93.84 Venugopalan[55 ] AD/HC sMRI+电子病历+基因数据 共220 十折交叉验证 AE,CNN,随机森林 88 Zeng[46 ] sMCI/pMCI sMRI 92 AD 训练集70% PCA,DBN(多任务) 87.78 Spasov[43 ] sMCI/pMCI sMRI 192 AD 十折交叉验证 CNN(多任务) 86 Liu[48 ] AD/HC sMRI 97 AD 五折交叉验证 V-Net,DenseNet 88.9 Abuhmed[15 ] AD/MCI/HC PET+sMRI+神经心理学数据+神经病理学数据+认知评分 共1371 十折交叉验证 BiLSTM,随机森林 84.95
10.11938/cjmr20223013.T0003 表3 可解释性方法及AD相关脑区 ...
Binary classification of Alzheimer’s disease using sMRI imaging modality and deep learning
2
2020
... Li等[34 ] 使用由文献[35 ]提供的3DSeg-8数据集作为预训练的源数据集,将权重迁移学习至ResNet-200,使AD、MCI和HC三分类准确率达到83%,而不使用迁移学习的准确率仅为76%.孔伶旭等[36 ] 使用ImageNet数据集迁移学习到Mobilenet网络的瓶颈层,然后将fMRI数据提取出来的ROI时间序列输入到网络中对网络顶层进行训练,最终网络对于EMCI与HC的分类准确率达到了73.67%.Bin等[37 ] 分别构建了平衡与不平衡数据集,同时在2D CNN、基于ImageNet迁移学习的Xception和Inception-v3网络中进行训练,最终发现带有迁移学习策略的Inception-v3在平衡数据集上性能最高,AD与HC分类准确率达到了99.45%. ...
... Performance comparison of different models
Table 2 第一作者 分类任务 数据模态 数据集 测试方法 分类模型 准确率% 郁松[21 ] AD/HC sMRI 1015 HC 训练集60% 3D ResNet-101 97.425 Parmar[24 ] AD/HC fMRI 30 AD 训练集60% 3D CNN 94.58 Bi[19 ] AD/HC fMRI 118 AD 五折交叉验证 RNN,ELM 91.3 (AUC) Yi̇Ği̇T[20 ] AD/HC sMRI 训练集 测试集 / 2D CNN 83 Punjabi[33 ] AD/HC PET 共1299 / 3D CNN 85.15 Zhang[49 ] AD/HC sMRI 200 AD 五折交叉验证 3D ResAttNet 91.3 Qiu[52 ] AD/HC sMRI+性别+年龄+MMSE / 训练集60% FCN 96.8 Deng[28 ] AD/HC DTI+sMRI 98 AD 训练集60% CNN 90.00 Marzban[29 ] AD/HC DTI+sMRI 115 AD 十折交叉验证 CNN 93.5 Kang[30 ] EMCI/HC DTI+sMRI 50 HC 训练集80% VGG-16,LASSO 94.2 Kwak[54 ] AD/HC sMRI 110 AD 五折交叉验证 DenseNet 93.75 Ju[16 ] MCI/HC fMRI 91 MCI 十折交叉验证 AE 86.47 Baydargil[17 ] AD/MCI/HC PET 141 AD 训练集80% CAE 98.67 Guan[51 ] AD/HC sMRI 384 AD 训练集 90% ResNet、pABN 90.7 Li[34 ] AD/MCI/HC MRI 237 AD 训练集65% ResNet-200 83 Basaia[40 ] AD/HC sMRI 294 AD 训练集90% 3D FCN(迁移学习) 99.2 孔伶旭[36 ] EMCI/HC fMRI 32 HC 五折交叉验证 Mobilenet(迁移学习) 73.67 Bin[37 ] AD/HC sMRI 100 AD 五折交叉验证 Inception-v3 99.45 Massalimova[27 ] AD/MCI/HC DTI 训练集 测试集 / ResNet-18(迁移学习) 97 Mehmood[38 ] AD/HC sMRI 75 AD 训练集64% VGG-19(迁移学习) 95.33 Raju[50 ] 非常轻度痴呆/轻度痴呆/中度痴呆/HC(四分类) sMRI 1013非常轻度痴呆 334非常轻度痴呆 VGG-16(迁移学习) 99 Lian[39 ] AD/HC sMRI 数据集1 数据集2 两个数据集间 H-FCN(迁移学习) 90.3 Oh[18 ] AD/HC sMRI 198 AD 五折交叉验证 3D CNN,ICAE 88.6 金祝新[53 ] AD/MCI sMRI 267 AD 训练集90% 3D CNN(迁移学习) 94.6 曾安[42 ] AD/HC sMRI 137 AD 五折交叉验证 2D CNN(集成学习) 81 Bi[22 ] AD/HC sMRI 243 AD / PCANet,k -means 89.15 Choi[41 ] AD/HC sMRI 715 AD 训练集60% VGG-16,GoogLeNet,AlexNet(集成学习) 93.84 Venugopalan[55 ] AD/HC sMRI+电子病历+基因数据 共220 十折交叉验证 AE,CNN,随机森林 88 Zeng[46 ] sMCI/pMCI sMRI 92 AD 训练集70% PCA,DBN(多任务) 87.78 Spasov[43 ] sMCI/pMCI sMRI 192 AD 十折交叉验证 CNN(多任务) 86 Liu[48 ] AD/HC sMRI 97 AD 五折交叉验证 V-Net,DenseNet 88.9 Abuhmed[15 ] AD/MCI/HC PET+sMRI+神经心理学数据+神经病理学数据+认知评分 共1371 十折交叉验证 BiLSTM,随机森林 84.95
10.11938/cjmr20223013.T0003 表3 可解释性方法及AD相关脑区 ...
A transfer learning approach for early diagnosis of Alzheimer’s disease on MRI images
2
2021
... 有很多研究者将迁移学习进一步应用到分类难度更大的不同类型MCI分类问题上,并且取得了不错的成果.Mehmood等[38 ] 将基于ImageNet数据集训练的权重迁移学习至VGG-19的前14层卷积层,使AD与HC分类准确率达到95.33%,EMCI与LMCI分类准确率达到83.72%.Lian等[39 ] 提出了一个分步提取斑块级和区域级特征的分层全卷积神经网络(hierarchical fully convolutional network,H-FCN)对AD与HC进行分类,并且将网络的权重迁移学习至sMCI与pMCI分类任务中,达到了80.9%的准确率.Basaia等[40 ] 使用FCN对AD与HC进行分类,并且将AD与HC分类网络的权重迁移学习到sMCI与pMCI分类网络中,在ADNI数据集中进行训练,分别在ADNI与独立测试集中进行测试,实验结果表明模型具有良好的泛化能力,在独立测试集中准确率仅从75.1%下降到了74.9%. ...
... Performance comparison of different models
Table 2 第一作者 分类任务 数据模态 数据集 测试方法 分类模型 准确率% 郁松[21 ] AD/HC sMRI 1015 HC 训练集60% 3D ResNet-101 97.425 Parmar[24 ] AD/HC fMRI 30 AD 训练集60% 3D CNN 94.58 Bi[19 ] AD/HC fMRI 118 AD 五折交叉验证 RNN,ELM 91.3 (AUC) Yi̇Ği̇T[20 ] AD/HC sMRI 训练集 测试集 / 2D CNN 83 Punjabi[33 ] AD/HC PET 共1299 / 3D CNN 85.15 Zhang[49 ] AD/HC sMRI 200 AD 五折交叉验证 3D ResAttNet 91.3 Qiu[52 ] AD/HC sMRI+性别+年龄+MMSE / 训练集60% FCN 96.8 Deng[28 ] AD/HC DTI+sMRI 98 AD 训练集60% CNN 90.00 Marzban[29 ] AD/HC DTI+sMRI 115 AD 十折交叉验证 CNN 93.5 Kang[30 ] EMCI/HC DTI+sMRI 50 HC 训练集80% VGG-16,LASSO 94.2 Kwak[54 ] AD/HC sMRI 110 AD 五折交叉验证 DenseNet 93.75 Ju[16 ] MCI/HC fMRI 91 MCI 十折交叉验证 AE 86.47 Baydargil[17 ] AD/MCI/HC PET 141 AD 训练集80% CAE 98.67 Guan[51 ] AD/HC sMRI 384 AD 训练集 90% ResNet、pABN 90.7 Li[34 ] AD/MCI/HC MRI 237 AD 训练集65% ResNet-200 83 Basaia[40 ] AD/HC sMRI 294 AD 训练集90% 3D FCN(迁移学习) 99.2 孔伶旭[36 ] EMCI/HC fMRI 32 HC 五折交叉验证 Mobilenet(迁移学习) 73.67 Bin[37 ] AD/HC sMRI 100 AD 五折交叉验证 Inception-v3 99.45 Massalimova[27 ] AD/MCI/HC DTI 训练集 测试集 / ResNet-18(迁移学习) 97 Mehmood[38 ] AD/HC sMRI 75 AD 训练集64% VGG-19(迁移学习) 95.33 Raju[50 ] 非常轻度痴呆/轻度痴呆/中度痴呆/HC(四分类) sMRI 1013非常轻度痴呆 334非常轻度痴呆 VGG-16(迁移学习) 99 Lian[39 ] AD/HC sMRI 数据集1 数据集2 两个数据集间 H-FCN(迁移学习) 90.3 Oh[18 ] AD/HC sMRI 198 AD 五折交叉验证 3D CNN,ICAE 88.6 金祝新[53 ] AD/MCI sMRI 267 AD 训练集90% 3D CNN(迁移学习) 94.6 曾安[42 ] AD/HC sMRI 137 AD 五折交叉验证 2D CNN(集成学习) 81 Bi[22 ] AD/HC sMRI 243 AD / PCANet,k -means 89.15 Choi[41 ] AD/HC sMRI 715 AD 训练集60% VGG-16,GoogLeNet,AlexNet(集成学习) 93.84 Venugopalan[55 ] AD/HC sMRI+电子病历+基因数据 共220 十折交叉验证 AE,CNN,随机森林 88 Zeng[46 ] sMCI/pMCI sMRI 92 AD 训练集70% PCA,DBN(多任务) 87.78 Spasov[43 ] sMCI/pMCI sMRI 192 AD 十折交叉验证 CNN(多任务) 86 Liu[48 ] AD/HC sMRI 97 AD 五折交叉验证 V-Net,DenseNet 88.9 Abuhmed[15 ] AD/MCI/HC PET+sMRI+神经心理学数据+神经病理学数据+认知评分 共1371 十折交叉验证 BiLSTM,随机森林 84.95
10.11938/cjmr20223013.T0003 表3 可解释性方法及AD相关脑区 ...
Hierarchical fully convolutional network for joint atrophy localization and Alzheimer’s disease diagnosis using structural MRI
3
2020
... 有很多研究者将迁移学习进一步应用到分类难度更大的不同类型MCI分类问题上,并且取得了不错的成果.Mehmood等[38 ] 将基于ImageNet数据集训练的权重迁移学习至VGG-19的前14层卷积层,使AD与HC分类准确率达到95.33%,EMCI与LMCI分类准确率达到83.72%.Lian等[39 ] 提出了一个分步提取斑块级和区域级特征的分层全卷积神经网络(hierarchical fully convolutional network,H-FCN)对AD与HC进行分类,并且将网络的权重迁移学习至sMCI与pMCI分类任务中,达到了80.9%的准确率.Basaia等[40 ] 使用FCN对AD与HC进行分类,并且将AD与HC分类网络的权重迁移学习到sMCI与pMCI分类网络中,在ADNI数据集中进行训练,分别在ADNI与独立测试集中进行测试,实验结果表明模型具有良好的泛化能力,在独立测试集中准确率仅从75.1%下降到了74.9%. ...
... 以往基于体素的研究方法可能会因为特征维数过高和影像数据量不足导致过拟合,而基于ROI区域的方法不能精确覆盖到全部的病理部位,所以近年来有研究者基于斑块级区域训练网络模型,如上文中提到的Lian等[39 ] 将大脑随机分割为若干个斑块区域对网络进行训练.这样的方法也可以用于生成神经网络热力图以达到可视化的效果,Qiu等[52 ] 通过FCN生成每个斑块对于AD预测的概率热力图,直观地反应大脑各个斑块级区域对于AD分类的重要性,结果显示网络模型生成的结果与AD病理学尸检结果高度相关,即AD概率的增加与海马、中额叶、杏仁核和颞叶区域中淀粉样蛋白β 和τ 的高水平积累相关.在热力图方法中,海马、颞叶和杏仁核被多次提及,与之前的医学研究结果相吻合,增加了研究者对于深度学习网络的信心. ...
... Performance comparison of different models
Table 2 第一作者 分类任务 数据模态 数据集 测试方法 分类模型 准确率% 郁松[21 ] AD/HC sMRI 1015 HC 训练集60% 3D ResNet-101 97.425 Parmar[24 ] AD/HC fMRI 30 AD 训练集60% 3D CNN 94.58 Bi[19 ] AD/HC fMRI 118 AD 五折交叉验证 RNN,ELM 91.3 (AUC) Yi̇Ği̇T[20 ] AD/HC sMRI 训练集 测试集 / 2D CNN 83 Punjabi[33 ] AD/HC PET 共1299 / 3D CNN 85.15 Zhang[49 ] AD/HC sMRI 200 AD 五折交叉验证 3D ResAttNet 91.3 Qiu[52 ] AD/HC sMRI+性别+年龄+MMSE / 训练集60% FCN 96.8 Deng[28 ] AD/HC DTI+sMRI 98 AD 训练集60% CNN 90.00 Marzban[29 ] AD/HC DTI+sMRI 115 AD 十折交叉验证 CNN 93.5 Kang[30 ] EMCI/HC DTI+sMRI 50 HC 训练集80% VGG-16,LASSO 94.2 Kwak[54 ] AD/HC sMRI 110 AD 五折交叉验证 DenseNet 93.75 Ju[16 ] MCI/HC fMRI 91 MCI 十折交叉验证 AE 86.47 Baydargil[17 ] AD/MCI/HC PET 141 AD 训练集80% CAE 98.67 Guan[51 ] AD/HC sMRI 384 AD 训练集 90% ResNet、pABN 90.7 Li[34 ] AD/MCI/HC MRI 237 AD 训练集65% ResNet-200 83 Basaia[40 ] AD/HC sMRI 294 AD 训练集90% 3D FCN(迁移学习) 99.2 孔伶旭[36 ] EMCI/HC fMRI 32 HC 五折交叉验证 Mobilenet(迁移学习) 73.67 Bin[37 ] AD/HC sMRI 100 AD 五折交叉验证 Inception-v3 99.45 Massalimova[27 ] AD/MCI/HC DTI 训练集 测试集 / ResNet-18(迁移学习) 97 Mehmood[38 ] AD/HC sMRI 75 AD 训练集64% VGG-19(迁移学习) 95.33 Raju[50 ] 非常轻度痴呆/轻度痴呆/中度痴呆/HC(四分类) sMRI 1013非常轻度痴呆 334非常轻度痴呆 VGG-16(迁移学习) 99 Lian[39 ] AD/HC sMRI 数据集1 数据集2 两个数据集间 H-FCN(迁移学习) 90.3 Oh[18 ] AD/HC sMRI 198 AD 五折交叉验证 3D CNN,ICAE 88.6 金祝新[53 ] AD/MCI sMRI 267 AD 训练集90% 3D CNN(迁移学习) 94.6 曾安[42 ] AD/HC sMRI 137 AD 五折交叉验证 2D CNN(集成学习) 81 Bi[22 ] AD/HC sMRI 243 AD / PCANet,k -means 89.15 Choi[41 ] AD/HC sMRI 715 AD 训练集60% VGG-16,GoogLeNet,AlexNet(集成学习) 93.84 Venugopalan[55 ] AD/HC sMRI+电子病历+基因数据 共220 十折交叉验证 AE,CNN,随机森林 88 Zeng[46 ] sMCI/pMCI sMRI 92 AD 训练集70% PCA,DBN(多任务) 87.78 Spasov[43 ] sMCI/pMCI sMRI 192 AD 十折交叉验证 CNN(多任务) 86 Liu[48 ] AD/HC sMRI 97 AD 五折交叉验证 V-Net,DenseNet 88.9 Abuhmed[15 ] AD/MCI/HC PET+sMRI+神经心理学数据+神经病理学数据+认知评分 共1371 十折交叉验证 BiLSTM,随机森林 84.95
10.11938/cjmr20223013.T0003 表3 可解释性方法及AD相关脑区 ...
Automated classification of Alzheimer's disease and mild cognitive impairment using a single MRI and deep neural networks
2
2019
... 有很多研究者将迁移学习进一步应用到分类难度更大的不同类型MCI分类问题上,并且取得了不错的成果.Mehmood等[38 ] 将基于ImageNet数据集训练的权重迁移学习至VGG-19的前14层卷积层,使AD与HC分类准确率达到95.33%,EMCI与LMCI分类准确率达到83.72%.Lian等[39 ] 提出了一个分步提取斑块级和区域级特征的分层全卷积神经网络(hierarchical fully convolutional network,H-FCN)对AD与HC进行分类,并且将网络的权重迁移学习至sMCI与pMCI分类任务中,达到了80.9%的准确率.Basaia等[40 ] 使用FCN对AD与HC进行分类,并且将AD与HC分类网络的权重迁移学习到sMCI与pMCI分类网络中,在ADNI数据集中进行训练,分别在ADNI与独立测试集中进行测试,实验结果表明模型具有良好的泛化能力,在独立测试集中准确率仅从75.1%下降到了74.9%. ...
... Performance comparison of different models
Table 2 第一作者 分类任务 数据模态 数据集 测试方法 分类模型 准确率% 郁松[21 ] AD/HC sMRI 1015 HC 训练集60% 3D ResNet-101 97.425 Parmar[24 ] AD/HC fMRI 30 AD 训练集60% 3D CNN 94.58 Bi[19 ] AD/HC fMRI 118 AD 五折交叉验证 RNN,ELM 91.3 (AUC) Yi̇Ği̇T[20 ] AD/HC sMRI 训练集 测试集 / 2D CNN 83 Punjabi[33 ] AD/HC PET 共1299 / 3D CNN 85.15 Zhang[49 ] AD/HC sMRI 200 AD 五折交叉验证 3D ResAttNet 91.3 Qiu[52 ] AD/HC sMRI+性别+年龄+MMSE / 训练集60% FCN 96.8 Deng[28 ] AD/HC DTI+sMRI 98 AD 训练集60% CNN 90.00 Marzban[29 ] AD/HC DTI+sMRI 115 AD 十折交叉验证 CNN 93.5 Kang[30 ] EMCI/HC DTI+sMRI 50 HC 训练集80% VGG-16,LASSO 94.2 Kwak[54 ] AD/HC sMRI 110 AD 五折交叉验证 DenseNet 93.75 Ju[16 ] MCI/HC fMRI 91 MCI 十折交叉验证 AE 86.47 Baydargil[17 ] AD/MCI/HC PET 141 AD 训练集80% CAE 98.67 Guan[51 ] AD/HC sMRI 384 AD 训练集 90% ResNet、pABN 90.7 Li[34 ] AD/MCI/HC MRI 237 AD 训练集65% ResNet-200 83 Basaia[40 ] AD/HC sMRI 294 AD 训练集90% 3D FCN(迁移学习) 99.2 孔伶旭[36 ] EMCI/HC fMRI 32 HC 五折交叉验证 Mobilenet(迁移学习) 73.67 Bin[37 ] AD/HC sMRI 100 AD 五折交叉验证 Inception-v3 99.45 Massalimova[27 ] AD/MCI/HC DTI 训练集 测试集 / ResNet-18(迁移学习) 97 Mehmood[38 ] AD/HC sMRI 75 AD 训练集64% VGG-19(迁移学习) 95.33 Raju[50 ] 非常轻度痴呆/轻度痴呆/中度痴呆/HC(四分类) sMRI 1013非常轻度痴呆 334非常轻度痴呆 VGG-16(迁移学习) 99 Lian[39 ] AD/HC sMRI 数据集1 数据集2 两个数据集间 H-FCN(迁移学习) 90.3 Oh[18 ] AD/HC sMRI 198 AD 五折交叉验证 3D CNN,ICAE 88.6 金祝新[53 ] AD/MCI sMRI 267 AD 训练集90% 3D CNN(迁移学习) 94.6 曾安[42 ] AD/HC sMRI 137 AD 五折交叉验证 2D CNN(集成学习) 81 Bi[22 ] AD/HC sMRI 243 AD / PCANet,k -means 89.15 Choi[41 ] AD/HC sMRI 715 AD 训练集60% VGG-16,GoogLeNet,AlexNet(集成学习) 93.84 Venugopalan[55 ] AD/HC sMRI+电子病历+基因数据 共220 十折交叉验证 AE,CNN,随机森林 88 Zeng[46 ] sMCI/pMCI sMRI 92 AD 训练集70% PCA,DBN(多任务) 87.78 Spasov[43 ] sMCI/pMCI sMRI 192 AD 十折交叉验证 CNN(多任务) 86 Liu[48 ] AD/HC sMRI 97 AD 五折交叉验证 V-Net,DenseNet 88.9 Abuhmed[15 ] AD/MCI/HC PET+sMRI+神经心理学数据+神经病理学数据+认知评分 共1371 十折交叉验证 BiLSTM,随机森林 84.95
10.11938/cjmr20223013.T0003 表3 可解释性方法及AD相关脑区 ...
Combining of multiple deep networks via ensemble generalization loss, based on mri images, for Alzheimer’s disease classification
2
2020
... 集成学习方法在AD影像分类任务中可以起到信息互补的作用.如果直接使用3D sMRI数据,会大幅增加网络参数量,若只使用单投影2D切片,则会损失其他投影方向的信息,所以有研究者使用集成学习方法融合3个轴向投影的互补信息,并取得了不错的结果.Choi等[41 ] 将sMRI图像的矢状面、冠状面和轴面分别作为VGG-16、GoogLeNet和AlexNet 3个CNN的输入,生成9个个体分类器,并构建了一个损失函数训练9个个体分类器的权重值,最终分类器在AD和HC分类任务中的准确率达到了93.84%.曾安等[42 ] 使用sMRI图像的矢状面、冠状面和轴面分别训练40个、50个和33个CNN基分类器,选出测试效果最好的5个基分类器使用投票法集成单轴分类器,最后使用投票法将3个单轴分类器集成为最终分类器,使AD与HC的分类准确率达到81%,pMCI与sMCI的分类准确率达到62%. ...
... Performance comparison of different models
Table 2 第一作者 分类任务 数据模态 数据集 测试方法 分类模型 准确率% 郁松[21 ] AD/HC sMRI 1015 HC 训练集60% 3D ResNet-101 97.425 Parmar[24 ] AD/HC fMRI 30 AD 训练集60% 3D CNN 94.58 Bi[19 ] AD/HC fMRI 118 AD 五折交叉验证 RNN,ELM 91.3 (AUC) Yi̇Ği̇T[20 ] AD/HC sMRI 训练集 测试集 / 2D CNN 83 Punjabi[33 ] AD/HC PET 共1299 / 3D CNN 85.15 Zhang[49 ] AD/HC sMRI 200 AD 五折交叉验证 3D ResAttNet 91.3 Qiu[52 ] AD/HC sMRI+性别+年龄+MMSE / 训练集60% FCN 96.8 Deng[28 ] AD/HC DTI+sMRI 98 AD 训练集60% CNN 90.00 Marzban[29 ] AD/HC DTI+sMRI 115 AD 十折交叉验证 CNN 93.5 Kang[30 ] EMCI/HC DTI+sMRI 50 HC 训练集80% VGG-16,LASSO 94.2 Kwak[54 ] AD/HC sMRI 110 AD 五折交叉验证 DenseNet 93.75 Ju[16 ] MCI/HC fMRI 91 MCI 十折交叉验证 AE 86.47 Baydargil[17 ] AD/MCI/HC PET 141 AD 训练集80% CAE 98.67 Guan[51 ] AD/HC sMRI 384 AD 训练集 90% ResNet、pABN 90.7 Li[34 ] AD/MCI/HC MRI 237 AD 训练集65% ResNet-200 83 Basaia[40 ] AD/HC sMRI 294 AD 训练集90% 3D FCN(迁移学习) 99.2 孔伶旭[36 ] EMCI/HC fMRI 32 HC 五折交叉验证 Mobilenet(迁移学习) 73.67 Bin[37 ] AD/HC sMRI 100 AD 五折交叉验证 Inception-v3 99.45 Massalimova[27 ] AD/MCI/HC DTI 训练集 测试集 / ResNet-18(迁移学习) 97 Mehmood[38 ] AD/HC sMRI 75 AD 训练集64% VGG-19(迁移学习) 95.33 Raju[50 ] 非常轻度痴呆/轻度痴呆/中度痴呆/HC(四分类) sMRI 1013非常轻度痴呆 334非常轻度痴呆 VGG-16(迁移学习) 99 Lian[39 ] AD/HC sMRI 数据集1 数据集2 两个数据集间 H-FCN(迁移学习) 90.3 Oh[18 ] AD/HC sMRI 198 AD 五折交叉验证 3D CNN,ICAE 88.6 金祝新[53 ] AD/MCI sMRI 267 AD 训练集90% 3D CNN(迁移学习) 94.6 曾安[42 ] AD/HC sMRI 137 AD 五折交叉验证 2D CNN(集成学习) 81 Bi[22 ] AD/HC sMRI 243 AD / PCANet,k -means 89.15 Choi[41 ] AD/HC sMRI 715 AD 训练集60% VGG-16,GoogLeNet,AlexNet(集成学习) 93.84 Venugopalan[55 ] AD/HC sMRI+电子病历+基因数据 共220 十折交叉验证 AE,CNN,随机森林 88 Zeng[46 ] sMCI/pMCI sMRI 92 AD 训练集70% PCA,DBN(多任务) 87.78 Spasov[43 ] sMCI/pMCI sMRI 192 AD 十折交叉验证 CNN(多任务) 86 Liu[48 ] AD/HC sMRI 97 AD 五折交叉验证 V-Net,DenseNet 88.9 Abuhmed[15 ] AD/MCI/HC PET+sMRI+神经心理学数据+神经病理学数据+认知评分 共1371 十折交叉验证 BiLSTM,随机森林 84.95
10.11938/cjmr20223013.T0003 表3 可解释性方法及AD相关脑区 ...
基于卷积神经网络和集成学习的阿尔茨海默症早期诊断
2
2019
... 集成学习方法在AD影像分类任务中可以起到信息互补的作用.如果直接使用3D sMRI数据,会大幅增加网络参数量,若只使用单投影2D切片,则会损失其他投影方向的信息,所以有研究者使用集成学习方法融合3个轴向投影的互补信息,并取得了不错的结果.Choi等[41 ] 将sMRI图像的矢状面、冠状面和轴面分别作为VGG-16、GoogLeNet和AlexNet 3个CNN的输入,生成9个个体分类器,并构建了一个损失函数训练9个个体分类器的权重值,最终分类器在AD和HC分类任务中的准确率达到了93.84%.曾安等[42 ] 使用sMRI图像的矢状面、冠状面和轴面分别训练40个、50个和33个CNN基分类器,选出测试效果最好的5个基分类器使用投票法集成单轴分类器,最后使用投票法将3个单轴分类器集成为最终分类器,使AD与HC的分类准确率达到81%,pMCI与sMCI的分类准确率达到62%. ...
... Performance comparison of different models
Table 2 第一作者 分类任务 数据模态 数据集 测试方法 分类模型 准确率% 郁松[21 ] AD/HC sMRI 1015 HC 训练集60% 3D ResNet-101 97.425 Parmar[24 ] AD/HC fMRI 30 AD 训练集60% 3D CNN 94.58 Bi[19 ] AD/HC fMRI 118 AD 五折交叉验证 RNN,ELM 91.3 (AUC) Yi̇Ği̇T[20 ] AD/HC sMRI 训练集 测试集 / 2D CNN 83 Punjabi[33 ] AD/HC PET 共1299 / 3D CNN 85.15 Zhang[49 ] AD/HC sMRI 200 AD 五折交叉验证 3D ResAttNet 91.3 Qiu[52 ] AD/HC sMRI+性别+年龄+MMSE / 训练集60% FCN 96.8 Deng[28 ] AD/HC DTI+sMRI 98 AD 训练集60% CNN 90.00 Marzban[29 ] AD/HC DTI+sMRI 115 AD 十折交叉验证 CNN 93.5 Kang[30 ] EMCI/HC DTI+sMRI 50 HC 训练集80% VGG-16,LASSO 94.2 Kwak[54 ] AD/HC sMRI 110 AD 五折交叉验证 DenseNet 93.75 Ju[16 ] MCI/HC fMRI 91 MCI 十折交叉验证 AE 86.47 Baydargil[17 ] AD/MCI/HC PET 141 AD 训练集80% CAE 98.67 Guan[51 ] AD/HC sMRI 384 AD 训练集 90% ResNet、pABN 90.7 Li[34 ] AD/MCI/HC MRI 237 AD 训练集65% ResNet-200 83 Basaia[40 ] AD/HC sMRI 294 AD 训练集90% 3D FCN(迁移学习) 99.2 孔伶旭[36 ] EMCI/HC fMRI 32 HC 五折交叉验证 Mobilenet(迁移学习) 73.67 Bin[37 ] AD/HC sMRI 100 AD 五折交叉验证 Inception-v3 99.45 Massalimova[27 ] AD/MCI/HC DTI 训练集 测试集 / ResNet-18(迁移学习) 97 Mehmood[38 ] AD/HC sMRI 75 AD 训练集64% VGG-19(迁移学习) 95.33 Raju[50 ] 非常轻度痴呆/轻度痴呆/中度痴呆/HC(四分类) sMRI 1013非常轻度痴呆 334非常轻度痴呆 VGG-16(迁移学习) 99 Lian[39 ] AD/HC sMRI 数据集1 数据集2 两个数据集间 H-FCN(迁移学习) 90.3 Oh[18 ] AD/HC sMRI 198 AD 五折交叉验证 3D CNN,ICAE 88.6 金祝新[53 ] AD/MCI sMRI 267 AD 训练集90% 3D CNN(迁移学习) 94.6 曾安[42 ] AD/HC sMRI 137 AD 五折交叉验证 2D CNN(集成学习) 81 Bi[22 ] AD/HC sMRI 243 AD / PCANet,k -means 89.15 Choi[41 ] AD/HC sMRI 715 AD 训练集60% VGG-16,GoogLeNet,AlexNet(集成学习) 93.84 Venugopalan[55 ] AD/HC sMRI+电子病历+基因数据 共220 十折交叉验证 AE,CNN,随机森林 88 Zeng[46 ] sMCI/pMCI sMRI 92 AD 训练集70% PCA,DBN(多任务) 87.78 Spasov[43 ] sMCI/pMCI sMRI 192 AD 十折交叉验证 CNN(多任务) 86 Liu[48 ] AD/HC sMRI 97 AD 五折交叉验证 V-Net,DenseNet 88.9 Abuhmed[15 ] AD/MCI/HC PET+sMRI+神经心理学数据+神经病理学数据+认知评分 共1371 十折交叉验证 BiLSTM,随机森林 84.95
10.11938/cjmr20223013.T0003 表3 可解释性方法及AD相关脑区 ...
基于卷积神经网络和集成学习的阿尔茨海默症早期诊断
2
2019
... 集成学习方法在AD影像分类任务中可以起到信息互补的作用.如果直接使用3D sMRI数据,会大幅增加网络参数量,若只使用单投影2D切片,则会损失其他投影方向的信息,所以有研究者使用集成学习方法融合3个轴向投影的互补信息,并取得了不错的结果.Choi等[41 ] 将sMRI图像的矢状面、冠状面和轴面分别作为VGG-16、GoogLeNet和AlexNet 3个CNN的输入,生成9个个体分类器,并构建了一个损失函数训练9个个体分类器的权重值,最终分类器在AD和HC分类任务中的准确率达到了93.84%.曾安等[42 ] 使用sMRI图像的矢状面、冠状面和轴面分别训练40个、50个和33个CNN基分类器,选出测试效果最好的5个基分类器使用投票法集成单轴分类器,最后使用投票法将3个单轴分类器集成为最终分类器,使AD与HC的分类准确率达到81%,pMCI与sMCI的分类准确率达到62%. ...
... Performance comparison of different models
Table 2 第一作者 分类任务 数据模态 数据集 测试方法 分类模型 准确率% 郁松[21 ] AD/HC sMRI 1015 HC 训练集60% 3D ResNet-101 97.425 Parmar[24 ] AD/HC fMRI 30 AD 训练集60% 3D CNN 94.58 Bi[19 ] AD/HC fMRI 118 AD 五折交叉验证 RNN,ELM 91.3 (AUC) Yi̇Ği̇T[20 ] AD/HC sMRI 训练集 测试集 / 2D CNN 83 Punjabi[33 ] AD/HC PET 共1299 / 3D CNN 85.15 Zhang[49 ] AD/HC sMRI 200 AD 五折交叉验证 3D ResAttNet 91.3 Qiu[52 ] AD/HC sMRI+性别+年龄+MMSE / 训练集60% FCN 96.8 Deng[28 ] AD/HC DTI+sMRI 98 AD 训练集60% CNN 90.00 Marzban[29 ] AD/HC DTI+sMRI 115 AD 十折交叉验证 CNN 93.5 Kang[30 ] EMCI/HC DTI+sMRI 50 HC 训练集80% VGG-16,LASSO 94.2 Kwak[54 ] AD/HC sMRI 110 AD 五折交叉验证 DenseNet 93.75 Ju[16 ] MCI/HC fMRI 91 MCI 十折交叉验证 AE 86.47 Baydargil[17 ] AD/MCI/HC PET 141 AD 训练集80% CAE 98.67 Guan[51 ] AD/HC sMRI 384 AD 训练集 90% ResNet、pABN 90.7 Li[34 ] AD/MCI/HC MRI 237 AD 训练集65% ResNet-200 83 Basaia[40 ] AD/HC sMRI 294 AD 训练集90% 3D FCN(迁移学习) 99.2 孔伶旭[36 ] EMCI/HC fMRI 32 HC 五折交叉验证 Mobilenet(迁移学习) 73.67 Bin[37 ] AD/HC sMRI 100 AD 五折交叉验证 Inception-v3 99.45 Massalimova[27 ] AD/MCI/HC DTI 训练集 测试集 / ResNet-18(迁移学习) 97 Mehmood[38 ] AD/HC sMRI 75 AD 训练集64% VGG-19(迁移学习) 95.33 Raju[50 ] 非常轻度痴呆/轻度痴呆/中度痴呆/HC(四分类) sMRI 1013非常轻度痴呆 334非常轻度痴呆 VGG-16(迁移学习) 99 Lian[39 ] AD/HC sMRI 数据集1 数据集2 两个数据集间 H-FCN(迁移学习) 90.3 Oh[18 ] AD/HC sMRI 198 AD 五折交叉验证 3D CNN,ICAE 88.6 金祝新[53 ] AD/MCI sMRI 267 AD 训练集90% 3D CNN(迁移学习) 94.6 曾安[42 ] AD/HC sMRI 137 AD 五折交叉验证 2D CNN(集成学习) 81 Bi[22 ] AD/HC sMRI 243 AD / PCANet,k -means 89.15 Choi[41 ] AD/HC sMRI 715 AD 训练集60% VGG-16,GoogLeNet,AlexNet(集成学习) 93.84 Venugopalan[55 ] AD/HC sMRI+电子病历+基因数据 共220 十折交叉验证 AE,CNN,随机森林 88 Zeng[46 ] sMCI/pMCI sMRI 92 AD 训练集70% PCA,DBN(多任务) 87.78 Spasov[43 ] sMCI/pMCI sMRI 192 AD 十折交叉验证 CNN(多任务) 86 Liu[48 ] AD/HC sMRI 97 AD 五折交叉验证 V-Net,DenseNet 88.9 Abuhmed[15 ] AD/MCI/HC PET+sMRI+神经心理学数据+神经病理学数据+认知评分 共1371 十折交叉验证 BiLSTM,随机森林 84.95
10.11938/cjmr20223013.T0003 表3 可解释性方法及AD相关脑区 ...
A parameter-efficient deep learning approach to predict conversion from mild cognitive impairment to Alzheimer’s disease
2
2019
... 前文中提到为了解决sMCI与pMCI分类模型难以训练的问题,有研究者使用迁移学习的方法将AD与HC的分类模型的权重迁移学习到sMCI与pMCI的分类模型中,可见两个任务之间存在高度的相似性,因此,同样可以用于多任务学习.如Spasov等[43 ] 结合了MRI图像作为3D CNN的输入,并且在最后全连接的输出向量后拼接上人口学数据、心理认知评估测试和Rey听觉言语学习测试等临床数据,作为多模态数据的融合向量输入到另一个全连接层中,以上部分的网络权重由两个任务共享,最后使用全连接层联合AD与HC、sMCI与pMCI两个分类任务进行训练,该模型在sMCI与pMCI分类任务中的准确率达到86%. ...
... Performance comparison of different models
Table 2 第一作者 分类任务 数据模态 数据集 测试方法 分类模型 准确率% 郁松[21 ] AD/HC sMRI 1015 HC 训练集60% 3D ResNet-101 97.425 Parmar[24 ] AD/HC fMRI 30 AD 训练集60% 3D CNN 94.58 Bi[19 ] AD/HC fMRI 118 AD 五折交叉验证 RNN,ELM 91.3 (AUC) Yi̇Ği̇T[20 ] AD/HC sMRI 训练集 测试集 / 2D CNN 83 Punjabi[33 ] AD/HC PET 共1299 / 3D CNN 85.15 Zhang[49 ] AD/HC sMRI 200 AD 五折交叉验证 3D ResAttNet 91.3 Qiu[52 ] AD/HC sMRI+性别+年龄+MMSE / 训练集60% FCN 96.8 Deng[28 ] AD/HC DTI+sMRI 98 AD 训练集60% CNN 90.00 Marzban[29 ] AD/HC DTI+sMRI 115 AD 十折交叉验证 CNN 93.5 Kang[30 ] EMCI/HC DTI+sMRI 50 HC 训练集80% VGG-16,LASSO 94.2 Kwak[54 ] AD/HC sMRI 110 AD 五折交叉验证 DenseNet 93.75 Ju[16 ] MCI/HC fMRI 91 MCI 十折交叉验证 AE 86.47 Baydargil[17 ] AD/MCI/HC PET 141 AD 训练集80% CAE 98.67 Guan[51 ] AD/HC sMRI 384 AD 训练集 90% ResNet、pABN 90.7 Li[34 ] AD/MCI/HC MRI 237 AD 训练集65% ResNet-200 83 Basaia[40 ] AD/HC sMRI 294 AD 训练集90% 3D FCN(迁移学习) 99.2 孔伶旭[36 ] EMCI/HC fMRI 32 HC 五折交叉验证 Mobilenet(迁移学习) 73.67 Bin[37 ] AD/HC sMRI 100 AD 五折交叉验证 Inception-v3 99.45 Massalimova[27 ] AD/MCI/HC DTI 训练集 测试集 / ResNet-18(迁移学习) 97 Mehmood[38 ] AD/HC sMRI 75 AD 训练集64% VGG-19(迁移学习) 95.33 Raju[50 ] 非常轻度痴呆/轻度痴呆/中度痴呆/HC(四分类) sMRI 1013非常轻度痴呆 334非常轻度痴呆 VGG-16(迁移学习) 99 Lian[39 ] AD/HC sMRI 数据集1 数据集2 两个数据集间 H-FCN(迁移学习) 90.3 Oh[18 ] AD/HC sMRI 198 AD 五折交叉验证 3D CNN,ICAE 88.6 金祝新[53 ] AD/MCI sMRI 267 AD 训练集90% 3D CNN(迁移学习) 94.6 曾安[42 ] AD/HC sMRI 137 AD 五折交叉验证 2D CNN(集成学习) 81 Bi[22 ] AD/HC sMRI 243 AD / PCANet,k -means 89.15 Choi[41 ] AD/HC sMRI 715 AD 训练集60% VGG-16,GoogLeNet,AlexNet(集成学习) 93.84 Venugopalan[55 ] AD/HC sMRI+电子病历+基因数据 共220 十折交叉验证 AE,CNN,随机森林 88 Zeng[46 ] sMCI/pMCI sMRI 92 AD 训练集70% PCA,DBN(多任务) 87.78 Spasov[43 ] sMCI/pMCI sMRI 192 AD 十折交叉验证 CNN(多任务) 86 Liu[48 ] AD/HC sMRI 97 AD 五折交叉验证 V-Net,DenseNet 88.9 Abuhmed[15 ] AD/MCI/HC PET+sMRI+神经心理学数据+神经病理学数据+认知评分 共1371 十折交叉验证 BiLSTM,随机森林 84.95
10.11938/cjmr20223013.T0003 表3 可解释性方法及AD相关脑区 ...
Self-reported hearing loss, hearing aids, and cognitive decline in elderly adults: A 25-year study
1
2015
... 在对AD患者进行诊断时,认知评分量表的分数是重要的参考标准[44 ] ,也有很多AD影像分类的论文[45 ] 将MMSE得分作为AD、MCI和HC组的标签,所以对于认知评分的回归预测也是AD影像分类多任务学习的理想辅助任务.如Zeng等[46 ] 将MMSE和AD评定量表-认知分量表(Alzheimer’s disease assessment scale,ADAS-cog)的回归预测作为辅助任务,使用深度信念网络(deep belief network,DBN)作为网络主干,最终得到的网络在AD与HC的分类、pMCI与HC的分类和AD与sMCI的分类中的准确率均超过95%.Abuhmed等[15 ] 使用患者在基线,以及基线后6个月、12个月和18个月的PET、sMRI、神经心理学、神经病理学和认知评分多模态数据,利用BiLSTM对患者基线后48个月的CDR和MMSE等7个认知评分进行回归预测,将预测得到的7个认知评分和患者的年龄、性别等信息作为随机森林分类器的输入,最后对患者基线48个月之后的状态进行AD、MCI与HC三分类,准确率达到了84.95%. ...
MRI deep learning-based solution for Alzheimer’s disease prediction
1
2021
... 在对AD患者进行诊断时,认知评分量表的分数是重要的参考标准[44 ] ,也有很多AD影像分类的论文[45 ] 将MMSE得分作为AD、MCI和HC组的标签,所以对于认知评分的回归预测也是AD影像分类多任务学习的理想辅助任务.如Zeng等[46 ] 将MMSE和AD评定量表-认知分量表(Alzheimer’s disease assessment scale,ADAS-cog)的回归预测作为辅助任务,使用深度信念网络(deep belief network,DBN)作为网络主干,最终得到的网络在AD与HC的分类、pMCI与HC的分类和AD与sMCI的分类中的准确率均超过95%.Abuhmed等[15 ] 使用患者在基线,以及基线后6个月、12个月和18个月的PET、sMRI、神经心理学、神经病理学和认知评分多模态数据,利用BiLSTM对患者基线后48个月的CDR和MMSE等7个认知评分进行回归预测,将预测得到的7个认知评分和患者的年龄、性别等信息作为随机森林分类器的输入,最后对患者基线48个月之后的状态进行AD、MCI与HC三分类,准确率达到了84.95%. ...
A new deep belief network-based multi-task learning for diagnosis of Alzheimer’s disease
2
... 在对AD患者进行诊断时,认知评分量表的分数是重要的参考标准[44 ] ,也有很多AD影像分类的论文[45 ] 将MMSE得分作为AD、MCI和HC组的标签,所以对于认知评分的回归预测也是AD影像分类多任务学习的理想辅助任务.如Zeng等[46 ] 将MMSE和AD评定量表-认知分量表(Alzheimer’s disease assessment scale,ADAS-cog)的回归预测作为辅助任务,使用深度信念网络(deep belief network,DBN)作为网络主干,最终得到的网络在AD与HC的分类、pMCI与HC的分类和AD与sMCI的分类中的准确率均超过95%.Abuhmed等[15 ] 使用患者在基线,以及基线后6个月、12个月和18个月的PET、sMRI、神经心理学、神经病理学和认知评分多模态数据,利用BiLSTM对患者基线后48个月的CDR和MMSE等7个认知评分进行回归预测,将预测得到的7个认知评分和患者的年龄、性别等信息作为随机森林分类器的输入,最后对患者基线48个月之后的状态进行AD、MCI与HC三分类,准确率达到了84.95%. ...
... Performance comparison of different models
Table 2 第一作者 分类任务 数据模态 数据集 测试方法 分类模型 准确率% 郁松[21 ] AD/HC sMRI 1015 HC 训练集60% 3D ResNet-101 97.425 Parmar[24 ] AD/HC fMRI 30 AD 训练集60% 3D CNN 94.58 Bi[19 ] AD/HC fMRI 118 AD 五折交叉验证 RNN,ELM 91.3 (AUC) Yi̇Ği̇T[20 ] AD/HC sMRI 训练集 测试集 / 2D CNN 83 Punjabi[33 ] AD/HC PET 共1299 / 3D CNN 85.15 Zhang[49 ] AD/HC sMRI 200 AD 五折交叉验证 3D ResAttNet 91.3 Qiu[52 ] AD/HC sMRI+性别+年龄+MMSE / 训练集60% FCN 96.8 Deng[28 ] AD/HC DTI+sMRI 98 AD 训练集60% CNN 90.00 Marzban[29 ] AD/HC DTI+sMRI 115 AD 十折交叉验证 CNN 93.5 Kang[30 ] EMCI/HC DTI+sMRI 50 HC 训练集80% VGG-16,LASSO 94.2 Kwak[54 ] AD/HC sMRI 110 AD 五折交叉验证 DenseNet 93.75 Ju[16 ] MCI/HC fMRI 91 MCI 十折交叉验证 AE 86.47 Baydargil[17 ] AD/MCI/HC PET 141 AD 训练集80% CAE 98.67 Guan[51 ] AD/HC sMRI 384 AD 训练集 90% ResNet、pABN 90.7 Li[34 ] AD/MCI/HC MRI 237 AD 训练集65% ResNet-200 83 Basaia[40 ] AD/HC sMRI 294 AD 训练集90% 3D FCN(迁移学习) 99.2 孔伶旭[36 ] EMCI/HC fMRI 32 HC 五折交叉验证 Mobilenet(迁移学习) 73.67 Bin[37 ] AD/HC sMRI 100 AD 五折交叉验证 Inception-v3 99.45 Massalimova[27 ] AD/MCI/HC DTI 训练集 测试集 / ResNet-18(迁移学习) 97 Mehmood[38 ] AD/HC sMRI 75 AD 训练集64% VGG-19(迁移学习) 95.33 Raju[50 ] 非常轻度痴呆/轻度痴呆/中度痴呆/HC(四分类) sMRI 1013非常轻度痴呆 334非常轻度痴呆 VGG-16(迁移学习) 99 Lian[39 ] AD/HC sMRI 数据集1 数据集2 两个数据集间 H-FCN(迁移学习) 90.3 Oh[18 ] AD/HC sMRI 198 AD 五折交叉验证 3D CNN,ICAE 88.6 金祝新[53 ] AD/MCI sMRI 267 AD 训练集90% 3D CNN(迁移学习) 94.6 曾安[42 ] AD/HC sMRI 137 AD 五折交叉验证 2D CNN(集成学习) 81 Bi[22 ] AD/HC sMRI 243 AD / PCANet,k -means 89.15 Choi[41 ] AD/HC sMRI 715 AD 训练集60% VGG-16,GoogLeNet,AlexNet(集成学习) 93.84 Venugopalan[55 ] AD/HC sMRI+电子病历+基因数据 共220 十折交叉验证 AE,CNN,随机森林 88 Zeng[46 ] sMCI/pMCI sMRI 92 AD 训练集70% PCA,DBN(多任务) 87.78 Spasov[43 ] sMCI/pMCI sMRI 192 AD 十折交叉验证 CNN(多任务) 86 Liu[48 ] AD/HC sMRI 97 AD 五折交叉验证 V-Net,DenseNet 88.9 Abuhmed[15 ] AD/MCI/HC PET+sMRI+神经心理学数据+神经病理学数据+认知评分 共1371 十折交叉验证 BiLSTM,随机森林 84.95
10.11938/cjmr20223013.T0003 表3 可解释性方法及AD相关脑区 ...
Predict Alzheimer’s disease using hippocampus MRI data: a lightweight 3D deep convolutional network model with visual and global shape representations
1
2021
... 有多项医学研究表明,AD是与海马体脑区高度相关的神经退行性疾病,同时也有不少研究者[47 ] 在进行AD影像分类时将海马区域作为ROI.如Liu等[48 ] 采用带有Dense模块的V-Net网络结构对3D MRI图像进行海马区域的分割,在进行海马分割的同时将从上采样层中提取的特征输入到全连接层进行AD与HC的分类,又融合了基于海马体掩膜提取到的特征进行分类,最终使AD与HC分类准确率达到88.9%,MCI与HC分类准确率达到76.2%. ...
A multi-model deep convolutional neural network for automatic hippocampus segmentation and classification in Alzheimer's disease
2
2020
... 有多项医学研究表明,AD是与海马体脑区高度相关的神经退行性疾病,同时也有不少研究者[47 ] 在进行AD影像分类时将海马区域作为ROI.如Liu等[48 ] 采用带有Dense模块的V-Net网络结构对3D MRI图像进行海马区域的分割,在进行海马分割的同时将从上采样层中提取的特征输入到全连接层进行AD与HC的分类,又融合了基于海马体掩膜提取到的特征进行分类,最终使AD与HC分类准确率达到88.9%,MCI与HC分类准确率达到76.2%. ...
... Performance comparison of different models
Table 2 第一作者 分类任务 数据模态 数据集 测试方法 分类模型 准确率% 郁松[21 ] AD/HC sMRI 1015 HC 训练集60% 3D ResNet-101 97.425 Parmar[24 ] AD/HC fMRI 30 AD 训练集60% 3D CNN 94.58 Bi[19 ] AD/HC fMRI 118 AD 五折交叉验证 RNN,ELM 91.3 (AUC) Yi̇Ği̇T[20 ] AD/HC sMRI 训练集 测试集 / 2D CNN 83 Punjabi[33 ] AD/HC PET 共1299 / 3D CNN 85.15 Zhang[49 ] AD/HC sMRI 200 AD 五折交叉验证 3D ResAttNet 91.3 Qiu[52 ] AD/HC sMRI+性别+年龄+MMSE / 训练集60% FCN 96.8 Deng[28 ] AD/HC DTI+sMRI 98 AD 训练集60% CNN 90.00 Marzban[29 ] AD/HC DTI+sMRI 115 AD 十折交叉验证 CNN 93.5 Kang[30 ] EMCI/HC DTI+sMRI 50 HC 训练集80% VGG-16,LASSO 94.2 Kwak[54 ] AD/HC sMRI 110 AD 五折交叉验证 DenseNet 93.75 Ju[16 ] MCI/HC fMRI 91 MCI 十折交叉验证 AE 86.47 Baydargil[17 ] AD/MCI/HC PET 141 AD 训练集80% CAE 98.67 Guan[51 ] AD/HC sMRI 384 AD 训练集 90% ResNet、pABN 90.7 Li[34 ] AD/MCI/HC MRI 237 AD 训练集65% ResNet-200 83 Basaia[40 ] AD/HC sMRI 294 AD 训练集90% 3D FCN(迁移学习) 99.2 孔伶旭[36 ] EMCI/HC fMRI 32 HC 五折交叉验证 Mobilenet(迁移学习) 73.67 Bin[37 ] AD/HC sMRI 100 AD 五折交叉验证 Inception-v3 99.45 Massalimova[27 ] AD/MCI/HC DTI 训练集 测试集 / ResNet-18(迁移学习) 97 Mehmood[38 ] AD/HC sMRI 75 AD 训练集64% VGG-19(迁移学习) 95.33 Raju[50 ] 非常轻度痴呆/轻度痴呆/中度痴呆/HC(四分类) sMRI 1013非常轻度痴呆 334非常轻度痴呆 VGG-16(迁移学习) 99 Lian[39 ] AD/HC sMRI 数据集1 数据集2 两个数据集间 H-FCN(迁移学习) 90.3 Oh[18 ] AD/HC sMRI 198 AD 五折交叉验证 3D CNN,ICAE 88.6 金祝新[53 ] AD/MCI sMRI 267 AD 训练集90% 3D CNN(迁移学习) 94.6 曾安[42 ] AD/HC sMRI 137 AD 五折交叉验证 2D CNN(集成学习) 81 Bi[22 ] AD/HC sMRI 243 AD / PCANet,k -means 89.15 Choi[41 ] AD/HC sMRI 715 AD 训练集60% VGG-16,GoogLeNet,AlexNet(集成学习) 93.84 Venugopalan[55 ] AD/HC sMRI+电子病历+基因数据 共220 十折交叉验证 AE,CNN,随机森林 88 Zeng[46 ] sMCI/pMCI sMRI 92 AD 训练集70% PCA,DBN(多任务) 87.78 Spasov[43 ] sMCI/pMCI sMRI 192 AD 十折交叉验证 CNN(多任务) 86 Liu[48 ] AD/HC sMRI 97 AD 五折交叉验证 V-Net,DenseNet 88.9 Abuhmed[15 ] AD/MCI/HC PET+sMRI+神经心理学数据+神经病理学数据+认知评分 共1371 十折交叉验证 BiLSTM,随机森林 84.95
10.11938/cjmr20223013.T0003 表3 可解释性方法及AD相关脑区 ...
An explainable 3D residual self-attention deep neural network for joint atrophy localization and Alzheimer’s disease diagnosis using structural MRI
3
2022
... Zhang等[49 ] 使用Grad-CAM对带有注意力机制的ResNet网络(3D ResAttNet)在AD与HC分类任务上进行可视化,热力图中突出显示了海马体、侧脑室和大部分皮质区域.Raju等[50 ] 使用Grad-CAM方法对认知障碍分类网络进行可视化操作,发现海马体、杏仁核和顶叶区域得到了最大程度的激活.Oh等[18 ] 使用ICAE对AD影像进行分类,并且使用类显著映射(class saliency visualization,CSV)手段,发现pMCI与sMCI的分类任务更注重于左侧杏仁核、角回和楔前回,而AD与HC分类任务更注重内侧颞叶周围、左侧海马体.Guan等[51 ] 结合了ResNet与并行注意力增强双线性网络(parallel attention-augmented bilinear network,pABN),使用CAM发现AD与HC分类、pMCI与sMCI分类的重点区域基本相似,集中于海马体、杏仁核、脑室、额叶、颞下回、颞上沟、顶枕沟和外侧裂. ...
... Performance comparison of different models
Table 2 第一作者 分类任务 数据模态 数据集 测试方法 分类模型 准确率% 郁松[21 ] AD/HC sMRI 1015 HC 训练集60% 3D ResNet-101 97.425 Parmar[24 ] AD/HC fMRI 30 AD 训练集60% 3D CNN 94.58 Bi[19 ] AD/HC fMRI 118 AD 五折交叉验证 RNN,ELM 91.3 (AUC) Yi̇Ği̇T[20 ] AD/HC sMRI 训练集 测试集 / 2D CNN 83 Punjabi[33 ] AD/HC PET 共1299 / 3D CNN 85.15 Zhang[49 ] AD/HC sMRI 200 AD 五折交叉验证 3D ResAttNet 91.3 Qiu[52 ] AD/HC sMRI+性别+年龄+MMSE / 训练集60% FCN 96.8 Deng[28 ] AD/HC DTI+sMRI 98 AD 训练集60% CNN 90.00 Marzban[29 ] AD/HC DTI+sMRI 115 AD 十折交叉验证 CNN 93.5 Kang[30 ] EMCI/HC DTI+sMRI 50 HC 训练集80% VGG-16,LASSO 94.2 Kwak[54 ] AD/HC sMRI 110 AD 五折交叉验证 DenseNet 93.75 Ju[16 ] MCI/HC fMRI 91 MCI 十折交叉验证 AE 86.47 Baydargil[17 ] AD/MCI/HC PET 141 AD 训练集80% CAE 98.67 Guan[51 ] AD/HC sMRI 384 AD 训练集 90% ResNet、pABN 90.7 Li[34 ] AD/MCI/HC MRI 237 AD 训练集65% ResNet-200 83 Basaia[40 ] AD/HC sMRI 294 AD 训练集90% 3D FCN(迁移学习) 99.2 孔伶旭[36 ] EMCI/HC fMRI 32 HC 五折交叉验证 Mobilenet(迁移学习) 73.67 Bin[37 ] AD/HC sMRI 100 AD 五折交叉验证 Inception-v3 99.45 Massalimova[27 ] AD/MCI/HC DTI 训练集 测试集 / ResNet-18(迁移学习) 97 Mehmood[38 ] AD/HC sMRI 75 AD 训练集64% VGG-19(迁移学习) 95.33 Raju[50 ] 非常轻度痴呆/轻度痴呆/中度痴呆/HC(四分类) sMRI 1013非常轻度痴呆 334非常轻度痴呆 VGG-16(迁移学习) 99 Lian[39 ] AD/HC sMRI 数据集1 数据集2 两个数据集间 H-FCN(迁移学习) 90.3 Oh[18 ] AD/HC sMRI 198 AD 五折交叉验证 3D CNN,ICAE 88.6 金祝新[53 ] AD/MCI sMRI 267 AD 训练集90% 3D CNN(迁移学习) 94.6 曾安[42 ] AD/HC sMRI 137 AD 五折交叉验证 2D CNN(集成学习) 81 Bi[22 ] AD/HC sMRI 243 AD / PCANet,k -means 89.15 Choi[41 ] AD/HC sMRI 715 AD 训练集60% VGG-16,GoogLeNet,AlexNet(集成学习) 93.84 Venugopalan[55 ] AD/HC sMRI+电子病历+基因数据 共220 十折交叉验证 AE,CNN,随机森林 88 Zeng[46 ] sMCI/pMCI sMRI 92 AD 训练集70% PCA,DBN(多任务) 87.78 Spasov[43 ] sMCI/pMCI sMRI 192 AD 十折交叉验证 CNN(多任务) 86 Liu[48 ] AD/HC sMRI 97 AD 五折交叉验证 V-Net,DenseNet 88.9 Abuhmed[15 ] AD/MCI/HC PET+sMRI+神经心理学数据+神经病理学数据+认知评分 共1371 十折交叉验证 BiLSTM,随机森林 84.95
10.11938/cjmr20223013.T0003 表3 可解释性方法及AD相关脑区 ...
... Comparison of different interpretation methods and AD-related brain regions
Table 3 第一作者 任务 分类模型 准确率% 解释性方法 脑区 Guan[51 ] AD/HC ResNet、pABN 90.7 CAM 海马体、杏仁核、脑室、额叶、颞下回、颞上沟、顶枕沟、外侧裂 Zhang[49 ] AD/HC 3D-ResAttNet 91.3 Grad-CAM 海马体、侧脑室、大部分皮质 Raju[50 ] 轻度痴呆/非常轻度痴呆/中度痴呆/HC(四分类) VGG-16(迁移学习) 99 Grad-CAM 海马体、杏仁核、顶叶 Oh[18 ] AD/HC 3D-CNN,ICAE 86.6 CSV 内侧颞叶周围、左侧海马体 Qiu[52 ] AD/HC FCN 96.8 基于斑块生成热力图 海马、中额叶、杏仁核、颞叶 金祝新[53 ] AD/HC 3D-CNN(迁移学习) 90.9 输入添加遮挡块 内侧颞叶、海马体 Kwak[54 ] sMCI/pMCI DenseNet 73.90 输入添加遮挡块 海马、梭状回、颞下回、楔前叶 Venugopalan[55 ] AD/HC AE,CNN,随机森林 88 特征屏蔽 海马体、杏仁核 Shahamat[56 ] AD/HC 3D-CNN 85 遗传算法选取脑模板 左侧枕叶、左侧颞梭状皮层、右侧楔皮层、右额中回、右颞中回
对于AD影像学分类及其模型可解释性研究,未来有以下发展方向:(1)探索更多对AD分类有效的数据模态,以及更多基于多模态数据融合分类的方法;(2)针对各个医疗机构成像条件不同带来的数据差异,进行更多跨数据集的模型测试用于优化模型的泛化能力;(3)在AD与HC分类问题的基础上,改进不同类型MCI分类或多分类任务的准确度;(4)针对AD影像学分类相关任务的深度学习模型,进行更多的可解释性研究,并发现更多可靠的与AD疾病相关的脑区. ...
Deep learning based multilevel classification of Alzheimer’s disease using MRI scans
3
2021
... Zhang等[49 ] 使用Grad-CAM对带有注意力机制的ResNet网络(3D ResAttNet)在AD与HC分类任务上进行可视化,热力图中突出显示了海马体、侧脑室和大部分皮质区域.Raju等[50 ] 使用Grad-CAM方法对认知障碍分类网络进行可视化操作,发现海马体、杏仁核和顶叶区域得到了最大程度的激活.Oh等[18 ] 使用ICAE对AD影像进行分类,并且使用类显著映射(class saliency visualization,CSV)手段,发现pMCI与sMCI的分类任务更注重于左侧杏仁核、角回和楔前回,而AD与HC分类任务更注重内侧颞叶周围、左侧海马体.Guan等[51 ] 结合了ResNet与并行注意力增强双线性网络(parallel attention-augmented bilinear network,pABN),使用CAM发现AD与HC分类、pMCI与sMCI分类的重点区域基本相似,集中于海马体、杏仁核、脑室、额叶、颞下回、颞上沟、顶枕沟和外侧裂. ...
... Performance comparison of different models
Table 2 第一作者 分类任务 数据模态 数据集 测试方法 分类模型 准确率% 郁松[21 ] AD/HC sMRI 1015 HC 训练集60% 3D ResNet-101 97.425 Parmar[24 ] AD/HC fMRI 30 AD 训练集60% 3D CNN 94.58 Bi[19 ] AD/HC fMRI 118 AD 五折交叉验证 RNN,ELM 91.3 (AUC) Yi̇Ği̇T[20 ] AD/HC sMRI 训练集 测试集 / 2D CNN 83 Punjabi[33 ] AD/HC PET 共1299 / 3D CNN 85.15 Zhang[49 ] AD/HC sMRI 200 AD 五折交叉验证 3D ResAttNet 91.3 Qiu[52 ] AD/HC sMRI+性别+年龄+MMSE / 训练集60% FCN 96.8 Deng[28 ] AD/HC DTI+sMRI 98 AD 训练集60% CNN 90.00 Marzban[29 ] AD/HC DTI+sMRI 115 AD 十折交叉验证 CNN 93.5 Kang[30 ] EMCI/HC DTI+sMRI 50 HC 训练集80% VGG-16,LASSO 94.2 Kwak[54 ] AD/HC sMRI 110 AD 五折交叉验证 DenseNet 93.75 Ju[16 ] MCI/HC fMRI 91 MCI 十折交叉验证 AE 86.47 Baydargil[17 ] AD/MCI/HC PET 141 AD 训练集80% CAE 98.67 Guan[51 ] AD/HC sMRI 384 AD 训练集 90% ResNet、pABN 90.7 Li[34 ] AD/MCI/HC MRI 237 AD 训练集65% ResNet-200 83 Basaia[40 ] AD/HC sMRI 294 AD 训练集90% 3D FCN(迁移学习) 99.2 孔伶旭[36 ] EMCI/HC fMRI 32 HC 五折交叉验证 Mobilenet(迁移学习) 73.67 Bin[37 ] AD/HC sMRI 100 AD 五折交叉验证 Inception-v3 99.45 Massalimova[27 ] AD/MCI/HC DTI 训练集 测试集 / ResNet-18(迁移学习) 97 Mehmood[38 ] AD/HC sMRI 75 AD 训练集64% VGG-19(迁移学习) 95.33 Raju[50 ] 非常轻度痴呆/轻度痴呆/中度痴呆/HC(四分类) sMRI 1013非常轻度痴呆 334非常轻度痴呆 VGG-16(迁移学习) 99 Lian[39 ] AD/HC sMRI 数据集1 数据集2 两个数据集间 H-FCN(迁移学习) 90.3 Oh[18 ] AD/HC sMRI 198 AD 五折交叉验证 3D CNN,ICAE 88.6 金祝新[53 ] AD/MCI sMRI 267 AD 训练集90% 3D CNN(迁移学习) 94.6 曾安[42 ] AD/HC sMRI 137 AD 五折交叉验证 2D CNN(集成学习) 81 Bi[22 ] AD/HC sMRI 243 AD / PCANet,k -means 89.15 Choi[41 ] AD/HC sMRI 715 AD 训练集60% VGG-16,GoogLeNet,AlexNet(集成学习) 93.84 Venugopalan[55 ] AD/HC sMRI+电子病历+基因数据 共220 十折交叉验证 AE,CNN,随机森林 88 Zeng[46 ] sMCI/pMCI sMRI 92 AD 训练集70% PCA,DBN(多任务) 87.78 Spasov[43 ] sMCI/pMCI sMRI 192 AD 十折交叉验证 CNN(多任务) 86 Liu[48 ] AD/HC sMRI 97 AD 五折交叉验证 V-Net,DenseNet 88.9 Abuhmed[15 ] AD/MCI/HC PET+sMRI+神经心理学数据+神经病理学数据+认知评分 共1371 十折交叉验证 BiLSTM,随机森林 84.95
10.11938/cjmr20223013.T0003 表3 可解释性方法及AD相关脑区 ...
... Comparison of different interpretation methods and AD-related brain regions
Table 3 第一作者 任务 分类模型 准确率% 解释性方法 脑区 Guan[51 ] AD/HC ResNet、pABN 90.7 CAM 海马体、杏仁核、脑室、额叶、颞下回、颞上沟、顶枕沟、外侧裂 Zhang[49 ] AD/HC 3D-ResAttNet 91.3 Grad-CAM 海马体、侧脑室、大部分皮质 Raju[50 ] 轻度痴呆/非常轻度痴呆/中度痴呆/HC(四分类) VGG-16(迁移学习) 99 Grad-CAM 海马体、杏仁核、顶叶 Oh[18 ] AD/HC 3D-CNN,ICAE 86.6 CSV 内侧颞叶周围、左侧海马体 Qiu[52 ] AD/HC FCN 96.8 基于斑块生成热力图 海马、中额叶、杏仁核、颞叶 金祝新[53 ] AD/HC 3D-CNN(迁移学习) 90.9 输入添加遮挡块 内侧颞叶、海马体 Kwak[54 ] sMCI/pMCI DenseNet 73.90 输入添加遮挡块 海马、梭状回、颞下回、楔前叶 Venugopalan[55 ] AD/HC AE,CNN,随机森林 88 特征屏蔽 海马体、杏仁核 Shahamat[56 ] AD/HC 3D-CNN 85 遗传算法选取脑模板 左侧枕叶、左侧颞梭状皮层、右侧楔皮层、右额中回、右颞中回
对于AD影像学分类及其模型可解释性研究,未来有以下发展方向:(1)探索更多对AD分类有效的数据模态,以及更多基于多模态数据融合分类的方法;(2)针对各个医疗机构成像条件不同带来的数据差异,进行更多跨数据集的模型测试用于优化模型的泛化能力;(3)在AD与HC分类问题的基础上,改进不同类型MCI分类或多分类任务的准确度;(4)针对AD影像学分类相关任务的深度学习模型,进行更多的可解释性研究,并发现更多可靠的与AD疾病相关的脑区. ...
A parallel attention-augmented bilinear network for early magnetic resonance imaging-based diagnosis of Alzheimer’s disease
3
2022
... Zhang等[49 ] 使用Grad-CAM对带有注意力机制的ResNet网络(3D ResAttNet)在AD与HC分类任务上进行可视化,热力图中突出显示了海马体、侧脑室和大部分皮质区域.Raju等[50 ] 使用Grad-CAM方法对认知障碍分类网络进行可视化操作,发现海马体、杏仁核和顶叶区域得到了最大程度的激活.Oh等[18 ] 使用ICAE对AD影像进行分类,并且使用类显著映射(class saliency visualization,CSV)手段,发现pMCI与sMCI的分类任务更注重于左侧杏仁核、角回和楔前回,而AD与HC分类任务更注重内侧颞叶周围、左侧海马体.Guan等[51 ] 结合了ResNet与并行注意力增强双线性网络(parallel attention-augmented bilinear network,pABN),使用CAM发现AD与HC分类、pMCI与sMCI分类的重点区域基本相似,集中于海马体、杏仁核、脑室、额叶、颞下回、颞上沟、顶枕沟和外侧裂. ...
... Performance comparison of different models
Table 2 第一作者 分类任务 数据模态 数据集 测试方法 分类模型 准确率% 郁松[21 ] AD/HC sMRI 1015 HC 训练集60% 3D ResNet-101 97.425 Parmar[24 ] AD/HC fMRI 30 AD 训练集60% 3D CNN 94.58 Bi[19 ] AD/HC fMRI 118 AD 五折交叉验证 RNN,ELM 91.3 (AUC) Yi̇Ği̇T[20 ] AD/HC sMRI 训练集 测试集 / 2D CNN 83 Punjabi[33 ] AD/HC PET 共1299 / 3D CNN 85.15 Zhang[49 ] AD/HC sMRI 200 AD 五折交叉验证 3D ResAttNet 91.3 Qiu[52 ] AD/HC sMRI+性别+年龄+MMSE / 训练集60% FCN 96.8 Deng[28 ] AD/HC DTI+sMRI 98 AD 训练集60% CNN 90.00 Marzban[29 ] AD/HC DTI+sMRI 115 AD 十折交叉验证 CNN 93.5 Kang[30 ] EMCI/HC DTI+sMRI 50 HC 训练集80% VGG-16,LASSO 94.2 Kwak[54 ] AD/HC sMRI 110 AD 五折交叉验证 DenseNet 93.75 Ju[16 ] MCI/HC fMRI 91 MCI 十折交叉验证 AE 86.47 Baydargil[17 ] AD/MCI/HC PET 141 AD 训练集80% CAE 98.67 Guan[51 ] AD/HC sMRI 384 AD 训练集 90% ResNet、pABN 90.7 Li[34 ] AD/MCI/HC MRI 237 AD 训练集65% ResNet-200 83 Basaia[40 ] AD/HC sMRI 294 AD 训练集90% 3D FCN(迁移学习) 99.2 孔伶旭[36 ] EMCI/HC fMRI 32 HC 五折交叉验证 Mobilenet(迁移学习) 73.67 Bin[37 ] AD/HC sMRI 100 AD 五折交叉验证 Inception-v3 99.45 Massalimova[27 ] AD/MCI/HC DTI 训练集 测试集 / ResNet-18(迁移学习) 97 Mehmood[38 ] AD/HC sMRI 75 AD 训练集64% VGG-19(迁移学习) 95.33 Raju[50 ] 非常轻度痴呆/轻度痴呆/中度痴呆/HC(四分类) sMRI 1013非常轻度痴呆 334非常轻度痴呆 VGG-16(迁移学习) 99 Lian[39 ] AD/HC sMRI 数据集1 数据集2 两个数据集间 H-FCN(迁移学习) 90.3 Oh[18 ] AD/HC sMRI 198 AD 五折交叉验证 3D CNN,ICAE 88.6 金祝新[53 ] AD/MCI sMRI 267 AD 训练集90% 3D CNN(迁移学习) 94.6 曾安[42 ] AD/HC sMRI 137 AD 五折交叉验证 2D CNN(集成学习) 81 Bi[22 ] AD/HC sMRI 243 AD / PCANet,k -means 89.15 Choi[41 ] AD/HC sMRI 715 AD 训练集60% VGG-16,GoogLeNet,AlexNet(集成学习) 93.84 Venugopalan[55 ] AD/HC sMRI+电子病历+基因数据 共220 十折交叉验证 AE,CNN,随机森林 88 Zeng[46 ] sMCI/pMCI sMRI 92 AD 训练集70% PCA,DBN(多任务) 87.78 Spasov[43 ] sMCI/pMCI sMRI 192 AD 十折交叉验证 CNN(多任务) 86 Liu[48 ] AD/HC sMRI 97 AD 五折交叉验证 V-Net,DenseNet 88.9 Abuhmed[15 ] AD/MCI/HC PET+sMRI+神经心理学数据+神经病理学数据+认知评分 共1371 十折交叉验证 BiLSTM,随机森林 84.95
10.11938/cjmr20223013.T0003 表3 可解释性方法及AD相关脑区 ...
... Comparison of different interpretation methods and AD-related brain regions
Table 3 第一作者 任务 分类模型 准确率% 解释性方法 脑区 Guan[51 ] AD/HC ResNet、pABN 90.7 CAM 海马体、杏仁核、脑室、额叶、颞下回、颞上沟、顶枕沟、外侧裂 Zhang[49 ] AD/HC 3D-ResAttNet 91.3 Grad-CAM 海马体、侧脑室、大部分皮质 Raju[50 ] 轻度痴呆/非常轻度痴呆/中度痴呆/HC(四分类) VGG-16(迁移学习) 99 Grad-CAM 海马体、杏仁核、顶叶 Oh[18 ] AD/HC 3D-CNN,ICAE 86.6 CSV 内侧颞叶周围、左侧海马体 Qiu[52 ] AD/HC FCN 96.8 基于斑块生成热力图 海马、中额叶、杏仁核、颞叶 金祝新[53 ] AD/HC 3D-CNN(迁移学习) 90.9 输入添加遮挡块 内侧颞叶、海马体 Kwak[54 ] sMCI/pMCI DenseNet 73.90 输入添加遮挡块 海马、梭状回、颞下回、楔前叶 Venugopalan[55 ] AD/HC AE,CNN,随机森林 88 特征屏蔽 海马体、杏仁核 Shahamat[56 ] AD/HC 3D-CNN 85 遗传算法选取脑模板 左侧枕叶、左侧颞梭状皮层、右侧楔皮层、右额中回、右颞中回
对于AD影像学分类及其模型可解释性研究,未来有以下发展方向:(1)探索更多对AD分类有效的数据模态,以及更多基于多模态数据融合分类的方法;(2)针对各个医疗机构成像条件不同带来的数据差异,进行更多跨数据集的模型测试用于优化模型的泛化能力;(3)在AD与HC分类问题的基础上,改进不同类型MCI分类或多分类任务的准确度;(4)针对AD影像学分类相关任务的深度学习模型,进行更多的可解释性研究,并发现更多可靠的与AD疾病相关的脑区. ...
Development and validation of an interpretable deep learning framework for Alzheimer’s disease classification
3
2020
... 以往基于体素的研究方法可能会因为特征维数过高和影像数据量不足导致过拟合,而基于ROI区域的方法不能精确覆盖到全部的病理部位,所以近年来有研究者基于斑块级区域训练网络模型,如上文中提到的Lian等[39 ] 将大脑随机分割为若干个斑块区域对网络进行训练.这样的方法也可以用于生成神经网络热力图以达到可视化的效果,Qiu等[52 ] 通过FCN生成每个斑块对于AD预测的概率热力图,直观地反应大脑各个斑块级区域对于AD分类的重要性,结果显示网络模型生成的结果与AD病理学尸检结果高度相关,即AD概率的增加与海马、中额叶、杏仁核和颞叶区域中淀粉样蛋白β 和τ 的高水平积累相关.在热力图方法中,海马、颞叶和杏仁核被多次提及,与之前的医学研究结果相吻合,增加了研究者对于深度学习网络的信心. ...
... Performance comparison of different models
Table 2 第一作者 分类任务 数据模态 数据集 测试方法 分类模型 准确率% 郁松[21 ] AD/HC sMRI 1015 HC 训练集60% 3D ResNet-101 97.425 Parmar[24 ] AD/HC fMRI 30 AD 训练集60% 3D CNN 94.58 Bi[19 ] AD/HC fMRI 118 AD 五折交叉验证 RNN,ELM 91.3 (AUC) Yi̇Ği̇T[20 ] AD/HC sMRI 训练集 测试集 / 2D CNN 83 Punjabi[33 ] AD/HC PET 共1299 / 3D CNN 85.15 Zhang[49 ] AD/HC sMRI 200 AD 五折交叉验证 3D ResAttNet 91.3 Qiu[52 ] AD/HC sMRI+性别+年龄+MMSE / 训练集60% FCN 96.8 Deng[28 ] AD/HC DTI+sMRI 98 AD 训练集60% CNN 90.00 Marzban[29 ] AD/HC DTI+sMRI 115 AD 十折交叉验证 CNN 93.5 Kang[30 ] EMCI/HC DTI+sMRI 50 HC 训练集80% VGG-16,LASSO 94.2 Kwak[54 ] AD/HC sMRI 110 AD 五折交叉验证 DenseNet 93.75 Ju[16 ] MCI/HC fMRI 91 MCI 十折交叉验证 AE 86.47 Baydargil[17 ] AD/MCI/HC PET 141 AD 训练集80% CAE 98.67 Guan[51 ] AD/HC sMRI 384 AD 训练集 90% ResNet、pABN 90.7 Li[34 ] AD/MCI/HC MRI 237 AD 训练集65% ResNet-200 83 Basaia[40 ] AD/HC sMRI 294 AD 训练集90% 3D FCN(迁移学习) 99.2 孔伶旭[36 ] EMCI/HC fMRI 32 HC 五折交叉验证 Mobilenet(迁移学习) 73.67 Bin[37 ] AD/HC sMRI 100 AD 五折交叉验证 Inception-v3 99.45 Massalimova[27 ] AD/MCI/HC DTI 训练集 测试集 / ResNet-18(迁移学习) 97 Mehmood[38 ] AD/HC sMRI 75 AD 训练集64% VGG-19(迁移学习) 95.33 Raju[50 ] 非常轻度痴呆/轻度痴呆/中度痴呆/HC(四分类) sMRI 1013非常轻度痴呆 334非常轻度痴呆 VGG-16(迁移学习) 99 Lian[39 ] AD/HC sMRI 数据集1 数据集2 两个数据集间 H-FCN(迁移学习) 90.3 Oh[18 ] AD/HC sMRI 198 AD 五折交叉验证 3D CNN,ICAE 88.6 金祝新[53 ] AD/MCI sMRI 267 AD 训练集90% 3D CNN(迁移学习) 94.6 曾安[42 ] AD/HC sMRI 137 AD 五折交叉验证 2D CNN(集成学习) 81 Bi[22 ] AD/HC sMRI 243 AD / PCANet,k -means 89.15 Choi[41 ] AD/HC sMRI 715 AD 训练集60% VGG-16,GoogLeNet,AlexNet(集成学习) 93.84 Venugopalan[55 ] AD/HC sMRI+电子病历+基因数据 共220 十折交叉验证 AE,CNN,随机森林 88 Zeng[46 ] sMCI/pMCI sMRI 92 AD 训练集70% PCA,DBN(多任务) 87.78 Spasov[43 ] sMCI/pMCI sMRI 192 AD 十折交叉验证 CNN(多任务) 86 Liu[48 ] AD/HC sMRI 97 AD 五折交叉验证 V-Net,DenseNet 88.9 Abuhmed[15 ] AD/MCI/HC PET+sMRI+神经心理学数据+神经病理学数据+认知评分 共1371 十折交叉验证 BiLSTM,随机森林 84.95
10.11938/cjmr20223013.T0003 表3 可解释性方法及AD相关脑区 ...
... Comparison of different interpretation methods and AD-related brain regions
Table 3 第一作者 任务 分类模型 准确率% 解释性方法 脑区 Guan[51 ] AD/HC ResNet、pABN 90.7 CAM 海马体、杏仁核、脑室、额叶、颞下回、颞上沟、顶枕沟、外侧裂 Zhang[49 ] AD/HC 3D-ResAttNet 91.3 Grad-CAM 海马体、侧脑室、大部分皮质 Raju[50 ] 轻度痴呆/非常轻度痴呆/中度痴呆/HC(四分类) VGG-16(迁移学习) 99 Grad-CAM 海马体、杏仁核、顶叶 Oh[18 ] AD/HC 3D-CNN,ICAE 86.6 CSV 内侧颞叶周围、左侧海马体 Qiu[52 ] AD/HC FCN 96.8 基于斑块生成热力图 海马、中额叶、杏仁核、颞叶 金祝新[53 ] AD/HC 3D-CNN(迁移学习) 90.9 输入添加遮挡块 内侧颞叶、海马体 Kwak[54 ] sMCI/pMCI DenseNet 73.90 输入添加遮挡块 海马、梭状回、颞下回、楔前叶 Venugopalan[55 ] AD/HC AE,CNN,随机森林 88 特征屏蔽 海马体、杏仁核 Shahamat[56 ] AD/HC 3D-CNN 85 遗传算法选取脑模板 左侧枕叶、左侧颞梭状皮层、右侧楔皮层、右额中回、右颞中回
对于AD影像学分类及其模型可解释性研究,未来有以下发展方向:(1)探索更多对AD分类有效的数据模态,以及更多基于多模态数据融合分类的方法;(2)针对各个医疗机构成像条件不同带来的数据差异,进行更多跨数据集的模型测试用于优化模型的泛化能力;(3)在AD与HC分类问题的基础上,改进不同类型MCI分类或多分类任务的准确度;(4)针对AD影像学分类相关任务的深度学习模型,进行更多的可解释性研究,并发现更多可靠的与AD疾病相关的脑区. ...
3
2019
... 金祝新等[53 ] 使用遮挡块的方式对网络进行可视化操作,即使用黑色或灰色色块遮挡输入图像,如果目标类别概率因被遮挡了某区域而降低,则认为该区域与AD分类任务相关,结果显示内侧颞叶和海马区域与AD影像分类具有高相关性.Kwak等[54 ] 同样使用遮挡块的方法对基于DenseNet的sMCI与pMCI分类网络进行了可解释性分析,发现海马、梭状回、颞下回和楔前叶区域在sMCI与pMCI分类任务中起到重要作用.Venugopalan等[55 ] 使用屏蔽输入特征的方法对CNN进行可解释性研究,结果显示MRI图像中海马和杏仁核对分类贡献最大. ...
... Performance comparison of different models
Table 2 第一作者 分类任务 数据模态 数据集 测试方法 分类模型 准确率% 郁松[21 ] AD/HC sMRI 1015 HC 训练集60% 3D ResNet-101 97.425 Parmar[24 ] AD/HC fMRI 30 AD 训练集60% 3D CNN 94.58 Bi[19 ] AD/HC fMRI 118 AD 五折交叉验证 RNN,ELM 91.3 (AUC) Yi̇Ği̇T[20 ] AD/HC sMRI 训练集 测试集 / 2D CNN 83 Punjabi[33 ] AD/HC PET 共1299 / 3D CNN 85.15 Zhang[49 ] AD/HC sMRI 200 AD 五折交叉验证 3D ResAttNet 91.3 Qiu[52 ] AD/HC sMRI+性别+年龄+MMSE / 训练集60% FCN 96.8 Deng[28 ] AD/HC DTI+sMRI 98 AD 训练集60% CNN 90.00 Marzban[29 ] AD/HC DTI+sMRI 115 AD 十折交叉验证 CNN 93.5 Kang[30 ] EMCI/HC DTI+sMRI 50 HC 训练集80% VGG-16,LASSO 94.2 Kwak[54 ] AD/HC sMRI 110 AD 五折交叉验证 DenseNet 93.75 Ju[16 ] MCI/HC fMRI 91 MCI 十折交叉验证 AE 86.47 Baydargil[17 ] AD/MCI/HC PET 141 AD 训练集80% CAE 98.67 Guan[51 ] AD/HC sMRI 384 AD 训练集 90% ResNet、pABN 90.7 Li[34 ] AD/MCI/HC MRI 237 AD 训练集65% ResNet-200 83 Basaia[40 ] AD/HC sMRI 294 AD 训练集90% 3D FCN(迁移学习) 99.2 孔伶旭[36 ] EMCI/HC fMRI 32 HC 五折交叉验证 Mobilenet(迁移学习) 73.67 Bin[37 ] AD/HC sMRI 100 AD 五折交叉验证 Inception-v3 99.45 Massalimova[27 ] AD/MCI/HC DTI 训练集 测试集 / ResNet-18(迁移学习) 97 Mehmood[38 ] AD/HC sMRI 75 AD 训练集64% VGG-19(迁移学习) 95.33 Raju[50 ] 非常轻度痴呆/轻度痴呆/中度痴呆/HC(四分类) sMRI 1013非常轻度痴呆 334非常轻度痴呆 VGG-16(迁移学习) 99 Lian[39 ] AD/HC sMRI 数据集1 数据集2 两个数据集间 H-FCN(迁移学习) 90.3 Oh[18 ] AD/HC sMRI 198 AD 五折交叉验证 3D CNN,ICAE 88.6 金祝新[53 ] AD/MCI sMRI 267 AD 训练集90% 3D CNN(迁移学习) 94.6 曾安[42 ] AD/HC sMRI 137 AD 五折交叉验证 2D CNN(集成学习) 81 Bi[22 ] AD/HC sMRI 243 AD / PCANet,k -means 89.15 Choi[41 ] AD/HC sMRI 715 AD 训练集60% VGG-16,GoogLeNet,AlexNet(集成学习) 93.84 Venugopalan[55 ] AD/HC sMRI+电子病历+基因数据 共220 十折交叉验证 AE,CNN,随机森林 88 Zeng[46 ] sMCI/pMCI sMRI 92 AD 训练集70% PCA,DBN(多任务) 87.78 Spasov[43 ] sMCI/pMCI sMRI 192 AD 十折交叉验证 CNN(多任务) 86 Liu[48 ] AD/HC sMRI 97 AD 五折交叉验证 V-Net,DenseNet 88.9 Abuhmed[15 ] AD/MCI/HC PET+sMRI+神经心理学数据+神经病理学数据+认知评分 共1371 十折交叉验证 BiLSTM,随机森林 84.95
10.11938/cjmr20223013.T0003 表3 可解释性方法及AD相关脑区 ...
... Comparison of different interpretation methods and AD-related brain regions
Table 3 第一作者 任务 分类模型 准确率% 解释性方法 脑区 Guan[51 ] AD/HC ResNet、pABN 90.7 CAM 海马体、杏仁核、脑室、额叶、颞下回、颞上沟、顶枕沟、外侧裂 Zhang[49 ] AD/HC 3D-ResAttNet 91.3 Grad-CAM 海马体、侧脑室、大部分皮质 Raju[50 ] 轻度痴呆/非常轻度痴呆/中度痴呆/HC(四分类) VGG-16(迁移学习) 99 Grad-CAM 海马体、杏仁核、顶叶 Oh[18 ] AD/HC 3D-CNN,ICAE 86.6 CSV 内侧颞叶周围、左侧海马体 Qiu[52 ] AD/HC FCN 96.8 基于斑块生成热力图 海马、中额叶、杏仁核、颞叶 金祝新[53 ] AD/HC 3D-CNN(迁移学习) 90.9 输入添加遮挡块 内侧颞叶、海马体 Kwak[54 ] sMCI/pMCI DenseNet 73.90 输入添加遮挡块 海马、梭状回、颞下回、楔前叶 Venugopalan[55 ] AD/HC AE,CNN,随机森林 88 特征屏蔽 海马体、杏仁核 Shahamat[56 ] AD/HC 3D-CNN 85 遗传算法选取脑模板 左侧枕叶、左侧颞梭状皮层、右侧楔皮层、右额中回、右颞中回
对于AD影像学分类及其模型可解释性研究,未来有以下发展方向:(1)探索更多对AD分类有效的数据模态,以及更多基于多模态数据融合分类的方法;(2)针对各个医疗机构成像条件不同带来的数据差异,进行更多跨数据集的模型测试用于优化模型的泛化能力;(3)在AD与HC分类问题的基础上,改进不同类型MCI分类或多分类任务的准确度;(4)针对AD影像学分类相关任务的深度学习模型,进行更多的可解释性研究,并发现更多可靠的与AD疾病相关的脑区. ...
Identifying the regional substrates predictive of Alzheimer’s disease progression through a convolutional neural network model and occlusion
3
2022
... 金祝新等[53 ] 使用遮挡块的方式对网络进行可视化操作,即使用黑色或灰色色块遮挡输入图像,如果目标类别概率因被遮挡了某区域而降低,则认为该区域与AD分类任务相关,结果显示内侧颞叶和海马区域与AD影像分类具有高相关性.Kwak等[54 ] 同样使用遮挡块的方法对基于DenseNet的sMCI与pMCI分类网络进行了可解释性分析,发现海马、梭状回、颞下回和楔前叶区域在sMCI与pMCI分类任务中起到重要作用.Venugopalan等[55 ] 使用屏蔽输入特征的方法对CNN进行可解释性研究,结果显示MRI图像中海马和杏仁核对分类贡献最大. ...
... Performance comparison of different models
Table 2 第一作者 分类任务 数据模态 数据集 测试方法 分类模型 准确率% 郁松[21 ] AD/HC sMRI 1015 HC 训练集60% 3D ResNet-101 97.425 Parmar[24 ] AD/HC fMRI 30 AD 训练集60% 3D CNN 94.58 Bi[19 ] AD/HC fMRI 118 AD 五折交叉验证 RNN,ELM 91.3 (AUC) Yi̇Ği̇T[20 ] AD/HC sMRI 训练集 测试集 / 2D CNN 83 Punjabi[33 ] AD/HC PET 共1299 / 3D CNN 85.15 Zhang[49 ] AD/HC sMRI 200 AD 五折交叉验证 3D ResAttNet 91.3 Qiu[52 ] AD/HC sMRI+性别+年龄+MMSE / 训练集60% FCN 96.8 Deng[28 ] AD/HC DTI+sMRI 98 AD 训练集60% CNN 90.00 Marzban[29 ] AD/HC DTI+sMRI 115 AD 十折交叉验证 CNN 93.5 Kang[30 ] EMCI/HC DTI+sMRI 50 HC 训练集80% VGG-16,LASSO 94.2 Kwak[54 ] AD/HC sMRI 110 AD 五折交叉验证 DenseNet 93.75 Ju[16 ] MCI/HC fMRI 91 MCI 十折交叉验证 AE 86.47 Baydargil[17 ] AD/MCI/HC PET 141 AD 训练集80% CAE 98.67 Guan[51 ] AD/HC sMRI 384 AD 训练集 90% ResNet、pABN 90.7 Li[34 ] AD/MCI/HC MRI 237 AD 训练集65% ResNet-200 83 Basaia[40 ] AD/HC sMRI 294 AD 训练集90% 3D FCN(迁移学习) 99.2 孔伶旭[36 ] EMCI/HC fMRI 32 HC 五折交叉验证 Mobilenet(迁移学习) 73.67 Bin[37 ] AD/HC sMRI 100 AD 五折交叉验证 Inception-v3 99.45 Massalimova[27 ] AD/MCI/HC DTI 训练集 测试集 / ResNet-18(迁移学习) 97 Mehmood[38 ] AD/HC sMRI 75 AD 训练集64% VGG-19(迁移学习) 95.33 Raju[50 ] 非常轻度痴呆/轻度痴呆/中度痴呆/HC(四分类) sMRI 1013非常轻度痴呆 334非常轻度痴呆 VGG-16(迁移学习) 99 Lian[39 ] AD/HC sMRI 数据集1 数据集2 两个数据集间 H-FCN(迁移学习) 90.3 Oh[18 ] AD/HC sMRI 198 AD 五折交叉验证 3D CNN,ICAE 88.6 金祝新[53 ] AD/MCI sMRI 267 AD 训练集90% 3D CNN(迁移学习) 94.6 曾安[42 ] AD/HC sMRI 137 AD 五折交叉验证 2D CNN(集成学习) 81 Bi[22 ] AD/HC sMRI 243 AD / PCANet,k -means 89.15 Choi[41 ] AD/HC sMRI 715 AD 训练集60% VGG-16,GoogLeNet,AlexNet(集成学习) 93.84 Venugopalan[55 ] AD/HC sMRI+电子病历+基因数据 共220 十折交叉验证 AE,CNN,随机森林 88 Zeng[46 ] sMCI/pMCI sMRI 92 AD 训练集70% PCA,DBN(多任务) 87.78 Spasov[43 ] sMCI/pMCI sMRI 192 AD 十折交叉验证 CNN(多任务) 86 Liu[48 ] AD/HC sMRI 97 AD 五折交叉验证 V-Net,DenseNet 88.9 Abuhmed[15 ] AD/MCI/HC PET+sMRI+神经心理学数据+神经病理学数据+认知评分 共1371 十折交叉验证 BiLSTM,随机森林 84.95
10.11938/cjmr20223013.T0003 表3 可解释性方法及AD相关脑区 ...
... Comparison of different interpretation methods and AD-related brain regions
Table 3 第一作者 任务 分类模型 准确率% 解释性方法 脑区 Guan[51 ] AD/HC ResNet、pABN 90.7 CAM 海马体、杏仁核、脑室、额叶、颞下回、颞上沟、顶枕沟、外侧裂 Zhang[49 ] AD/HC 3D-ResAttNet 91.3 Grad-CAM 海马体、侧脑室、大部分皮质 Raju[50 ] 轻度痴呆/非常轻度痴呆/中度痴呆/HC(四分类) VGG-16(迁移学习) 99 Grad-CAM 海马体、杏仁核、顶叶 Oh[18 ] AD/HC 3D-CNN,ICAE 86.6 CSV 内侧颞叶周围、左侧海马体 Qiu[52 ] AD/HC FCN 96.8 基于斑块生成热力图 海马、中额叶、杏仁核、颞叶 金祝新[53 ] AD/HC 3D-CNN(迁移学习) 90.9 输入添加遮挡块 内侧颞叶、海马体 Kwak[54 ] sMCI/pMCI DenseNet 73.90 输入添加遮挡块 海马、梭状回、颞下回、楔前叶 Venugopalan[55 ] AD/HC AE,CNN,随机森林 88 特征屏蔽 海马体、杏仁核 Shahamat[56 ] AD/HC 3D-CNN 85 遗传算法选取脑模板 左侧枕叶、左侧颞梭状皮层、右侧楔皮层、右额中回、右颞中回
对于AD影像学分类及其模型可解释性研究,未来有以下发展方向:(1)探索更多对AD分类有效的数据模态,以及更多基于多模态数据融合分类的方法;(2)针对各个医疗机构成像条件不同带来的数据差异,进行更多跨数据集的模型测试用于优化模型的泛化能力;(3)在AD与HC分类问题的基础上,改进不同类型MCI分类或多分类任务的准确度;(4)针对AD影像学分类相关任务的深度学习模型,进行更多的可解释性研究,并发现更多可靠的与AD疾病相关的脑区. ...
Multimodal deep learning models for early detection of Alzheimer’s disease stage
3
2021
... 金祝新等[53 ] 使用遮挡块的方式对网络进行可视化操作,即使用黑色或灰色色块遮挡输入图像,如果目标类别概率因被遮挡了某区域而降低,则认为该区域与AD分类任务相关,结果显示内侧颞叶和海马区域与AD影像分类具有高相关性.Kwak等[54 ] 同样使用遮挡块的方法对基于DenseNet的sMCI与pMCI分类网络进行了可解释性分析,发现海马、梭状回、颞下回和楔前叶区域在sMCI与pMCI分类任务中起到重要作用.Venugopalan等[55 ] 使用屏蔽输入特征的方法对CNN进行可解释性研究,结果显示MRI图像中海马和杏仁核对分类贡献最大. ...
... Performance comparison of different models
Table 2 第一作者 分类任务 数据模态 数据集 测试方法 分类模型 准确率% 郁松[21 ] AD/HC sMRI 1015 HC 训练集60% 3D ResNet-101 97.425 Parmar[24 ] AD/HC fMRI 30 AD 训练集60% 3D CNN 94.58 Bi[19 ] AD/HC fMRI 118 AD 五折交叉验证 RNN,ELM 91.3 (AUC) Yi̇Ği̇T[20 ] AD/HC sMRI 训练集 测试集 / 2D CNN 83 Punjabi[33 ] AD/HC PET 共1299 / 3D CNN 85.15 Zhang[49 ] AD/HC sMRI 200 AD 五折交叉验证 3D ResAttNet 91.3 Qiu[52 ] AD/HC sMRI+性别+年龄+MMSE / 训练集60% FCN 96.8 Deng[28 ] AD/HC DTI+sMRI 98 AD 训练集60% CNN 90.00 Marzban[29 ] AD/HC DTI+sMRI 115 AD 十折交叉验证 CNN 93.5 Kang[30 ] EMCI/HC DTI+sMRI 50 HC 训练集80% VGG-16,LASSO 94.2 Kwak[54 ] AD/HC sMRI 110 AD 五折交叉验证 DenseNet 93.75 Ju[16 ] MCI/HC fMRI 91 MCI 十折交叉验证 AE 86.47 Baydargil[17 ] AD/MCI/HC PET 141 AD 训练集80% CAE 98.67 Guan[51 ] AD/HC sMRI 384 AD 训练集 90% ResNet、pABN 90.7 Li[34 ] AD/MCI/HC MRI 237 AD 训练集65% ResNet-200 83 Basaia[40 ] AD/HC sMRI 294 AD 训练集90% 3D FCN(迁移学习) 99.2 孔伶旭[36 ] EMCI/HC fMRI 32 HC 五折交叉验证 Mobilenet(迁移学习) 73.67 Bin[37 ] AD/HC sMRI 100 AD 五折交叉验证 Inception-v3 99.45 Massalimova[27 ] AD/MCI/HC DTI 训练集 测试集 / ResNet-18(迁移学习) 97 Mehmood[38 ] AD/HC sMRI 75 AD 训练集64% VGG-19(迁移学习) 95.33 Raju[50 ] 非常轻度痴呆/轻度痴呆/中度痴呆/HC(四分类) sMRI 1013非常轻度痴呆 334非常轻度痴呆 VGG-16(迁移学习) 99 Lian[39 ] AD/HC sMRI 数据集1 数据集2 两个数据集间 H-FCN(迁移学习) 90.3 Oh[18 ] AD/HC sMRI 198 AD 五折交叉验证 3D CNN,ICAE 88.6 金祝新[53 ] AD/MCI sMRI 267 AD 训练集90% 3D CNN(迁移学习) 94.6 曾安[42 ] AD/HC sMRI 137 AD 五折交叉验证 2D CNN(集成学习) 81 Bi[22 ] AD/HC sMRI 243 AD / PCANet,k -means 89.15 Choi[41 ] AD/HC sMRI 715 AD 训练集60% VGG-16,GoogLeNet,AlexNet(集成学习) 93.84 Venugopalan[55 ] AD/HC sMRI+电子病历+基因数据 共220 十折交叉验证 AE,CNN,随机森林 88 Zeng[46 ] sMCI/pMCI sMRI 92 AD 训练集70% PCA,DBN(多任务) 87.78 Spasov[43 ] sMCI/pMCI sMRI 192 AD 十折交叉验证 CNN(多任务) 86 Liu[48 ] AD/HC sMRI 97 AD 五折交叉验证 V-Net,DenseNet 88.9 Abuhmed[15 ] AD/MCI/HC PET+sMRI+神经心理学数据+神经病理学数据+认知评分 共1371 十折交叉验证 BiLSTM,随机森林 84.95
10.11938/cjmr20223013.T0003 表3 可解释性方法及AD相关脑区 ...
... Comparison of different interpretation methods and AD-related brain regions
Table 3 第一作者 任务 分类模型 准确率% 解释性方法 脑区 Guan[51 ] AD/HC ResNet、pABN 90.7 CAM 海马体、杏仁核、脑室、额叶、颞下回、颞上沟、顶枕沟、外侧裂 Zhang[49 ] AD/HC 3D-ResAttNet 91.3 Grad-CAM 海马体、侧脑室、大部分皮质 Raju[50 ] 轻度痴呆/非常轻度痴呆/中度痴呆/HC(四分类) VGG-16(迁移学习) 99 Grad-CAM 海马体、杏仁核、顶叶 Oh[18 ] AD/HC 3D-CNN,ICAE 86.6 CSV 内侧颞叶周围、左侧海马体 Qiu[52 ] AD/HC FCN 96.8 基于斑块生成热力图 海马、中额叶、杏仁核、颞叶 金祝新[53 ] AD/HC 3D-CNN(迁移学习) 90.9 输入添加遮挡块 内侧颞叶、海马体 Kwak[54 ] sMCI/pMCI DenseNet 73.90 输入添加遮挡块 海马、梭状回、颞下回、楔前叶 Venugopalan[55 ] AD/HC AE,CNN,随机森林 88 特征屏蔽 海马体、杏仁核 Shahamat[56 ] AD/HC 3D-CNN 85 遗传算法选取脑模板 左侧枕叶、左侧颞梭状皮层、右侧楔皮层、右额中回、右颞中回
对于AD影像学分类及其模型可解释性研究,未来有以下发展方向:(1)探索更多对AD分类有效的数据模态,以及更多基于多模态数据融合分类的方法;(2)针对各个医疗机构成像条件不同带来的数据差异,进行更多跨数据集的模型测试用于优化模型的泛化能力;(3)在AD与HC分类问题的基础上,改进不同类型MCI分类或多分类任务的准确度;(4)针对AD影像学分类相关任务的深度学习模型,进行更多的可解释性研究,并发现更多可靠的与AD疾病相关的脑区. ...
Brain MRI analysis using a deep learning based evolutionary approach
2
2020
... 以上的研究都是通过人工选取保留或剔除的特征,具有一定的随机性和主观性,而使用算法自动选取脑区则可以避免这样的问题.Shahamat等[56 ] 从哈佛牛津大脑图谱中获得96个大脑区域,将它们存储到与输入大小相同的3D矩阵中.基于这些脑区使用遗传算法随机生成掩膜,掩膜中可以包含一个或多个脑区,作为网络的输入.通过不断减小掩膜大小,最终选择了被认为对AD影像分类最具贡献的5个脑区,分别是左侧枕叶、左侧颞梭状皮层、右侧楔皮层、右额中回和右颞中回.输入消融实验法对于脑区的定位可以精确到人为定义的解剖模板,相比神经网络热力图可以定位到更细致的区域. ...
... Comparison of different interpretation methods and AD-related brain regions
Table 3 第一作者 任务 分类模型 准确率% 解释性方法 脑区 Guan[51 ] AD/HC ResNet、pABN 90.7 CAM 海马体、杏仁核、脑室、额叶、颞下回、颞上沟、顶枕沟、外侧裂 Zhang[49 ] AD/HC 3D-ResAttNet 91.3 Grad-CAM 海马体、侧脑室、大部分皮质 Raju[50 ] 轻度痴呆/非常轻度痴呆/中度痴呆/HC(四分类) VGG-16(迁移学习) 99 Grad-CAM 海马体、杏仁核、顶叶 Oh[18 ] AD/HC 3D-CNN,ICAE 86.6 CSV 内侧颞叶周围、左侧海马体 Qiu[52 ] AD/HC FCN 96.8 基于斑块生成热力图 海马、中额叶、杏仁核、颞叶 金祝新[53 ] AD/HC 3D-CNN(迁移学习) 90.9 输入添加遮挡块 内侧颞叶、海马体 Kwak[54 ] sMCI/pMCI DenseNet 73.90 输入添加遮挡块 海马、梭状回、颞下回、楔前叶 Venugopalan[55 ] AD/HC AE,CNN,随机森林 88 特征屏蔽 海马体、杏仁核 Shahamat[56 ] AD/HC 3D-CNN 85 遗传算法选取脑模板 左侧枕叶、左侧颞梭状皮层、右侧楔皮层、右额中回、右颞中回
对于AD影像学分类及其模型可解释性研究,未来有以下发展方向:(1)探索更多对AD分类有效的数据模态,以及更多基于多模态数据融合分类的方法;(2)针对各个医疗机构成像条件不同带来的数据差异,进行更多跨数据集的模型测试用于优化模型的泛化能力;(3)在AD与HC分类问题的基础上,改进不同类型MCI分类或多分类任务的准确度;(4)针对AD影像学分类相关任务的深度学习模型,进行更多的可解释性研究,并发现更多可靠的与AD疾病相关的脑区. ...
Predicting brain age using machine learning algorithms: A comprehensive evaluation
1
2022
... 近几年,大脑年龄的预测逐渐变成了研究的热点,而AD是一种与年龄高度相关的疾病,Beheshti等[57 ] 整合了15种脑龄预测模型,发现在AD组中的测试误差明显高于MCI组,MCI组明显高于HC组,可见患者患病程度越高,越会导致脑龄预测模型高估其大脑年龄.有些研究者针对脑龄预测与AD影像分类之间的关联性进行探索.如Bashyam等[58 ] 使用正常人的sMRI图像训练了一个包含有Inception与ResNet模块的CNN网络,在测试集上MAE为3.701,将脑龄预测模型的权重值迁移学习到AD与HC分类模型中,实验结果表明在小样本情况下,该迁移学习模型的性能远远高于基于ImageNet迁移学习的分类模型,可见脑龄预测与AD影像分类任务有着极高的相关性.探索脑龄预测任务与AD影像学分类任务的关联性或许也可以成为未来的一个研究方向. ...
MRI signatures of brain age and disease over the lifespan based on a deep brain network and 14 468 individuals worldwide
1
2020
... 近几年,大脑年龄的预测逐渐变成了研究的热点,而AD是一种与年龄高度相关的疾病,Beheshti等[57 ] 整合了15种脑龄预测模型,发现在AD组中的测试误差明显高于MCI组,MCI组明显高于HC组,可见患者患病程度越高,越会导致脑龄预测模型高估其大脑年龄.有些研究者针对脑龄预测与AD影像分类之间的关联性进行探索.如Bashyam等[58 ] 使用正常人的sMRI图像训练了一个包含有Inception与ResNet模块的CNN网络,在测试集上MAE为3.701,将脑龄预测模型的权重值迁移学习到AD与HC分类模型中,实验结果表明在小样本情况下,该迁移学习模型的性能远远高于基于ImageNet迁移学习的分类模型,可见脑龄预测与AD影像分类任务有着极高的相关性.探索脑龄预测任务与AD影像学分类任务的关联性或许也可以成为未来的一个研究方向. ...